![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW242_AGTCAA_L004_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 6313438 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 40 |
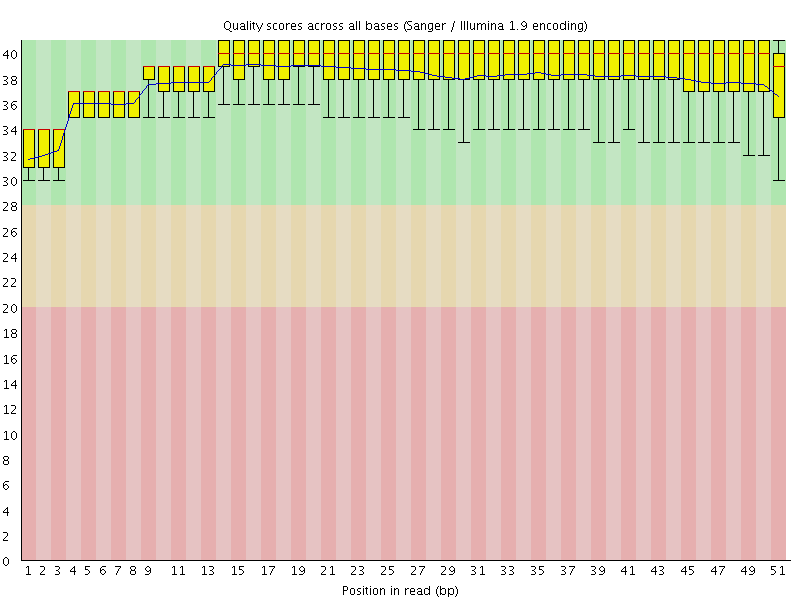
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
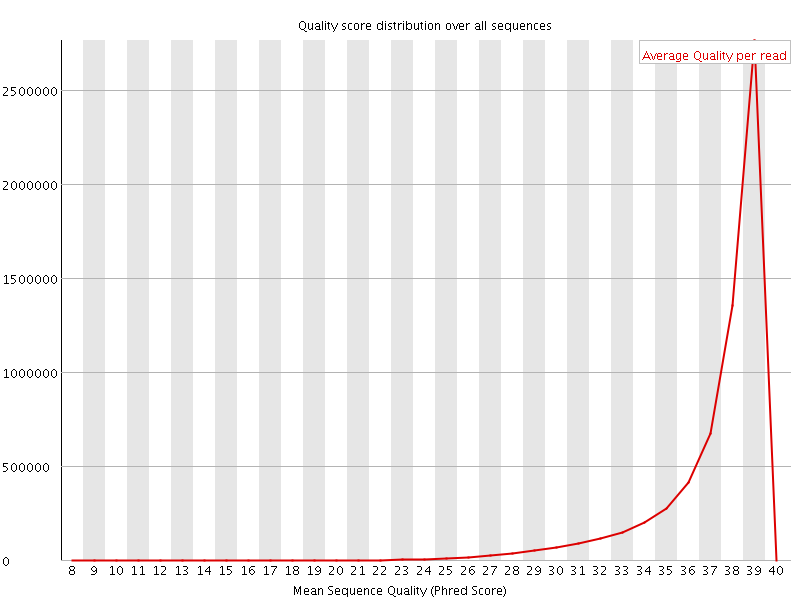
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
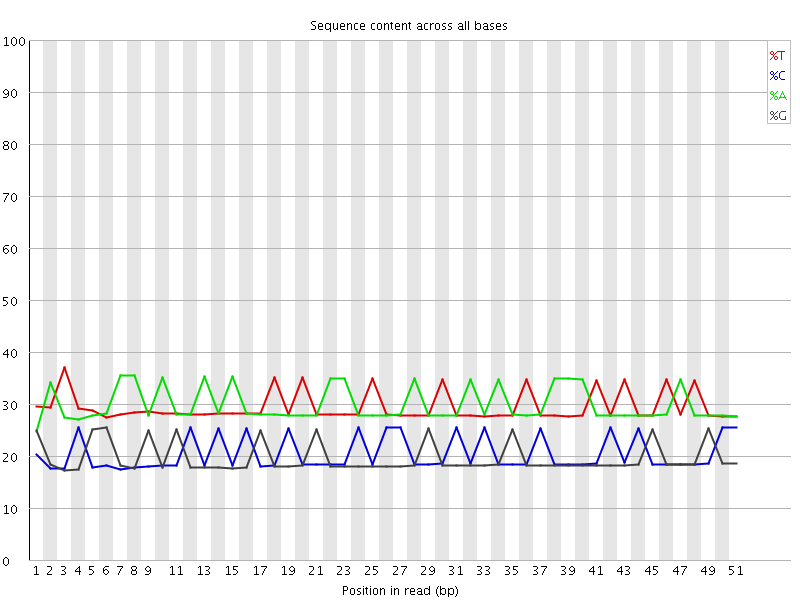
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
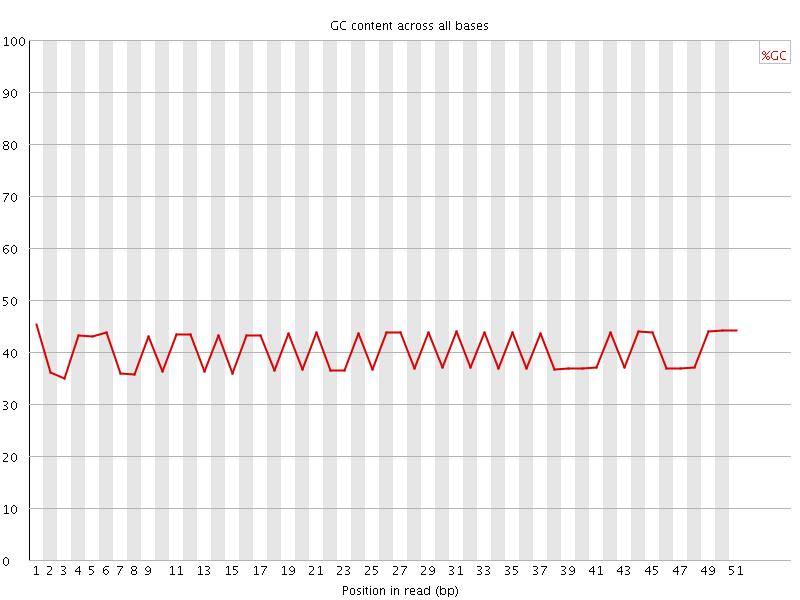
![[WARN]](Icons/warning.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
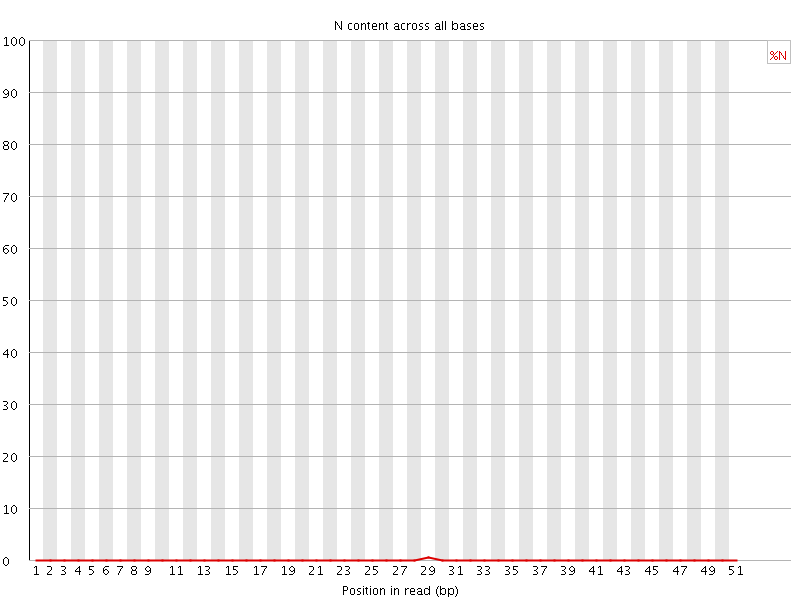
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
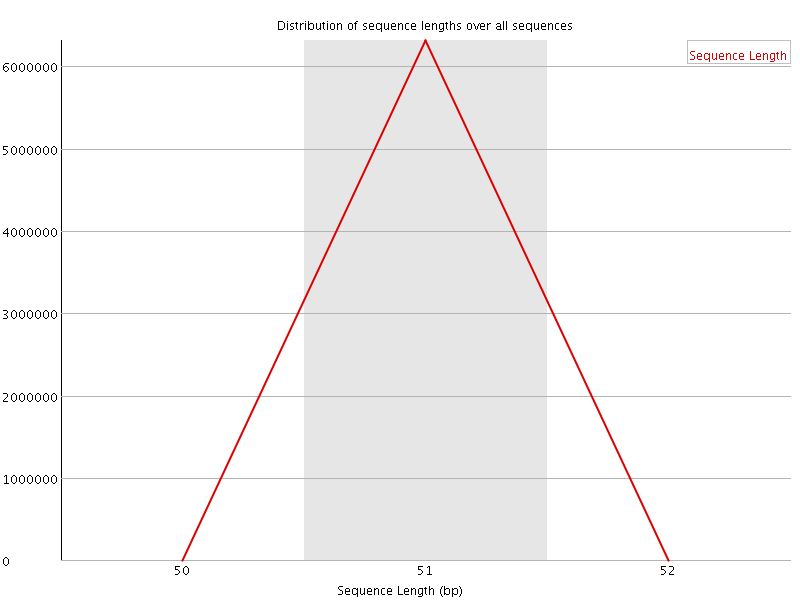
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACAGTCAAATCTCGTATGCC | 395980 | 6.272018510358382 | TruSeq Adapter, Index 8 (97% over 36bp) |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
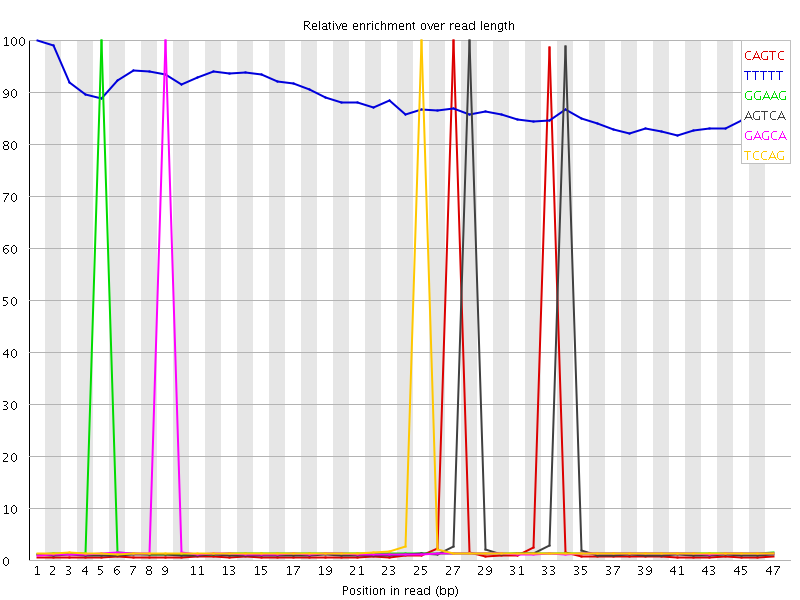
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CAGTC | 1059850 | 4.8002768 | 94.67202 | 27 |
| TTTTT | 2504545 | 3.74005 | 4.2361135 | 1 |
| GGAAG | 766025 | 3.6934004 | 104.479195 | 5 |
| AGTCA | 1138675 | 3.5279636 | 64.78312 | 28 |
| GAGCA | 745335 | 3.4528892 | 99.69417 | 9 |
| TCCAG | 760345 | 3.4437575 | 95.65545 | 25 |
| AAAAA | 2482615 | 3.3989382 | 3.6470342 | 12 |
| CGGAA | 726970 | 3.3678102 | 100.77523 | 4 |
| CTCCA | 767790 | 3.3412738 | 92.05169 | 24 |
| TCTCG | 677015 | 3.1200614 | 95.65407 | 41 |
| TCGGA | 659760 | 3.109998 | 102.18888 | 3 |
| AGAGC | 669780 | 3.1028686 | 99.41357 | 8 |
| CACAG | 690425 | 3.0732327 | 93.02651 | 31 |
| GTCTG | 639290 | 3.0663025 | 101.96817 | 17 |
| CCAGT | 665315 | 3.0133471 | 94.99306 | 26 |
| ATCGG | 637140 | 3.003371 | 100.70034 | 2 |
| GAAGA | 946310 | 2.9989347 | 69.02769 | 6 |
| AGCAC | 672315 | 2.992621 | 95.802925 | 10 |
| GCACA | 672010 | 2.9912634 | 95.62004 | 11 |
| CTGAA | 947335 | 2.9351335 | 66.71536 | 19 |
| CACAC | 681690 | 2.9155037 | 91.77244 | 12 |
| GATCG | 612770 | 2.8884952 | 99.77164 | 1 |
| ATGCC | 635970 | 2.8804376 | 93.10214 | 47 |
| CGTCT | 623955 | 2.8755312 | 98.05038 | 16 |
| GTCAC | 632435 | 2.864427 | 94.76534 | 29 |
| CTCGT | 615790 | 2.8379025 | 94.755295 | 42 |
| CACGT | 615520 | 2.7878156 | 96.57753 | 14 |
| ACTCC | 634805 | 2.7625487 | 91.959984 | 23 |
| ACGTC | 599905 | 2.717092 | 96.420616 | 15 |
| ACACG | 600605 | 2.6734242 | 94.98151 | 13 |
| TCTGA | 831570 | 2.6215975 | 67.67382 | 18 |
| TCAAA | 1276330 | 2.599186 | 43.03587 | 36 |
| AAGAG | 780655 | 2.4739604 | 68.42353 | 7 |
| TGAAC | 777950 | 2.410327 | 66.1008 | 20 |
| TCACA | 780775 | 2.3243306 | 62.34291 | 30 |
| CAAAT | 1117810 | 2.2763674 | 42.920128 | 37 |
| GAACT | 732890 | 2.2707176 | 65.90728 | 21 |
| ACAGT | 732495 | 2.2694933 | 64.40158 | 32 |
| GTCAA | 731950 | 2.2678049 | 64.12832 | 35 |
| ATCTC | 740795 | 2.2439492 | 63.13973 | 40 |
| AACTC | 748535 | 2.2283537 | 63.42518 | 22 |
| AAATC | 1093285 | 2.2264235 | 42.94078 | 38 |
| GTATG | 614750 | 2.0170562 | 67.321915 | 45 |
| TATGC | 628715 | 1.9820794 | 64.57752 | 46 |
| TCGTA | 621090 | 1.958041 | 64.78784 | 43 |
| CGTAT | 598470 | 1.8867294 | 64.64281 | 44 |
| AATCT | 849040 | 1.7593225 | 43.22505 | 39 |
| TGCCG | 234845 | 1.6182777 | 5.1103406 | 47 |