![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW234_ACAGTG_L004_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 8776965 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 37 |
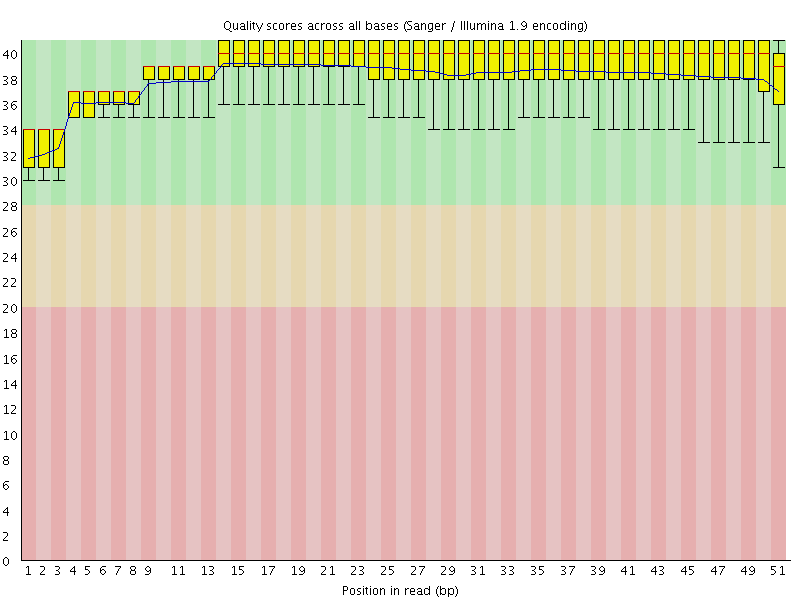
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
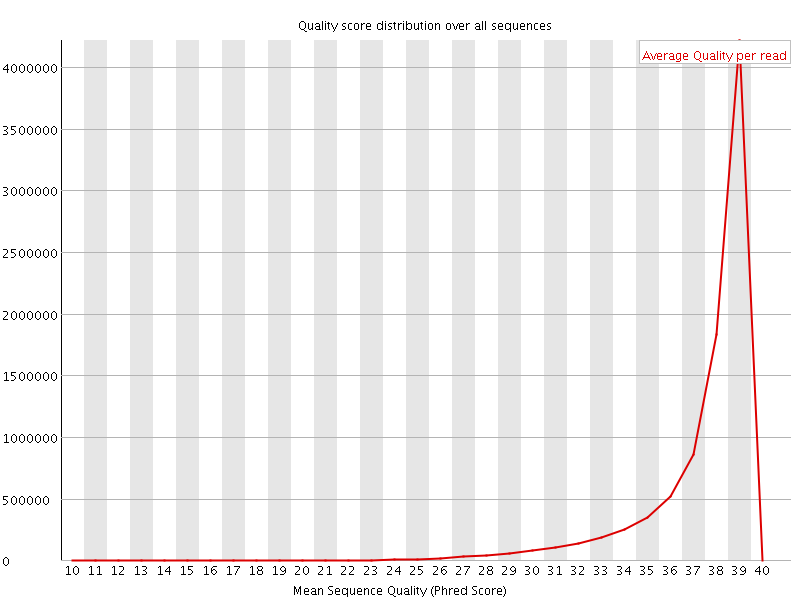
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
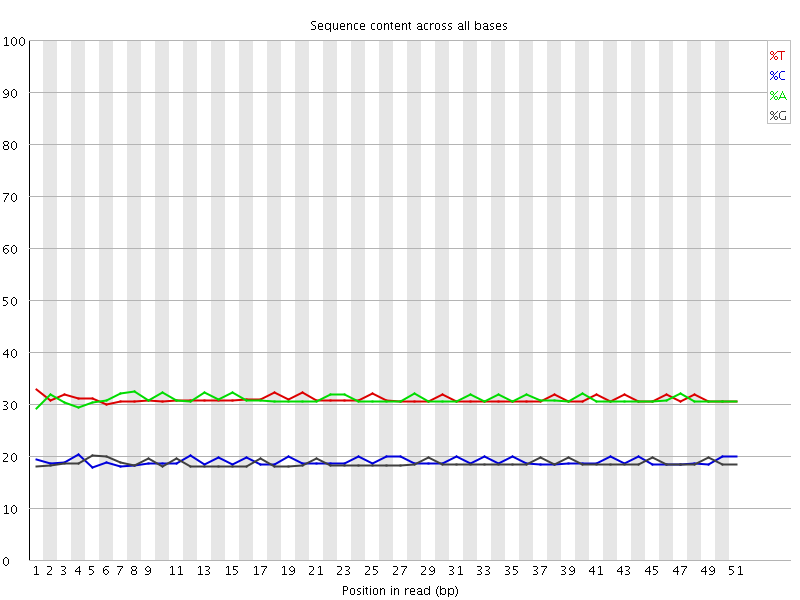
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
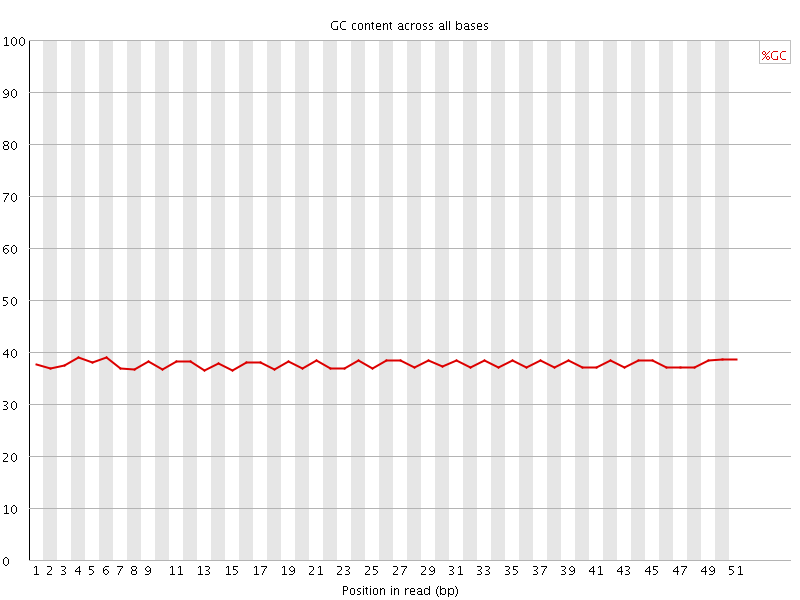
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
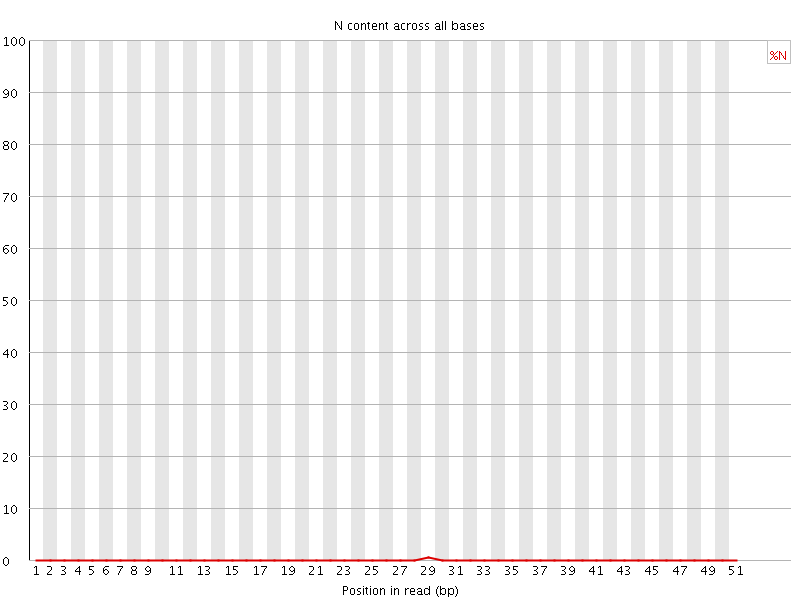
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
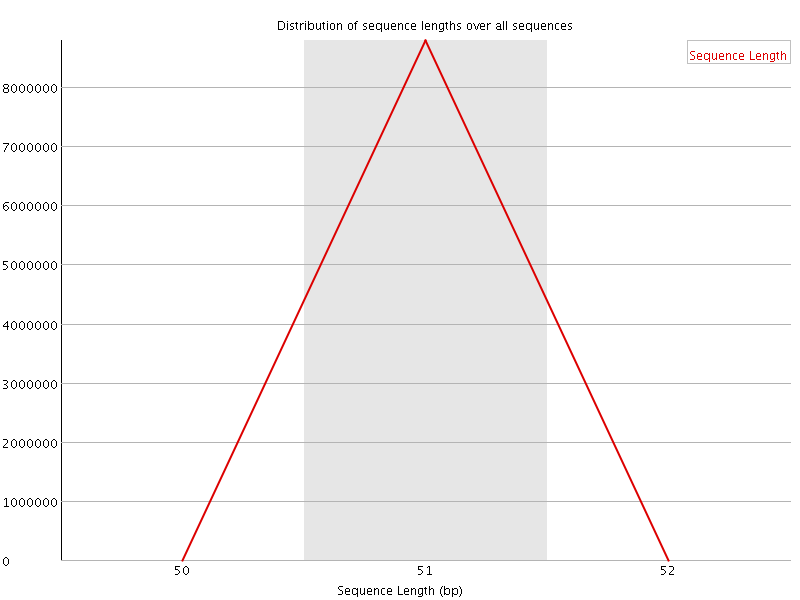
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACACAGTGATCTCGTATGCC | 107683 | 1.2268819574875827 | TruSeq Adapter, Index 5 (100% over 51bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content

| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 4238815 | 3.540963 | 3.7842376 | 15 |
| AAAAA | 4190730 | 3.5135098 | 3.6845748 | 13 |
| TTTTC | 2220325 | 3.0215316 | 3.3787098 | 1 |
| CTCCA | 598720 | 2.1637974 | 22.523823 | 24 |
| GAGCA | 570450 | 2.1356006 | 23.767235 | 9 |
| CGGAA | 563300 | 2.1088328 | 24.1144 | 4 |
| GGAAG | 549220 | 2.091925 | 24.330437 | 5 |
| TCCAG | 560595 | 2.0612915 | 22.748001 | 25 |
| CACAC | 568075 | 2.0545347 | 22.465601 | 12 |
| GAAGA | 889925 | 2.0466194 | 15.424171 | 6 |
| CTGAA | 901235 | 2.03568 | 15.038784 | 19 |
| TCGGA | 474585 | 1.7754217 | 23.707838 | 3 |
| TCTCG | 475830 | 1.7483448 | 22.39843 | 41 |
| TCTGA | 720850 | 1.627052 | 14.664605 | 18 |
| GCACA | 441780 | 1.6255904 | 22.89999 | 11 |
| CAGTG | 434510 | 1.6255012 | 22.463291 | 35 |
| AGAGC | 430330 | 1.6110315 | 23.294369 | 8 |
| CACAG | 422945 | 1.5562845 | 22.134377 | 33 |
| AGCAC | 422140 | 1.5533223 | 22.889624 | 10 |
| ATCGG | 403795 | 1.5105964 | 23.108866 | 2 |
| CCAGT | 407460 | 1.4982184 | 22.127697 | 26 |
| GTCAC | 391275 | 1.4387066 | 22.133818 | 29 |
| TGAAC | 635000 | 1.434317 | 14.404114 | 20 |
| CTCGT | 387610 | 1.4241974 | 22.081072 | 42 |
| TCACA | 637475 | 1.4152633 | 13.880475 | 30 |
| AAGAG | 605110 | 1.3916116 | 14.748179 | 7 |
| CACGT | 376120 | 1.3829823 | 22.585564 | 14 |
| GTCTG | 368560 | 1.377783 | 22.915806 | 17 |
| CGTCT | 373345 | 1.3717835 | 22.540043 | 16 |
| ACTCC | 377640 | 1.3648057 | 21.8602 | 23 |
| AGTGA | 574705 | 1.320729 | 14.108338 | 36 |
| ATGCC | 356135 | 1.309498 | 22.008303 | 47 |
| ACACG | 355140 | 1.3067865 | 22.536293 | 13 |
| GATCG | 343040 | 1.283312 | 22.688673 | 1 |
| ACACA | 577280 | 1.2825536 | 13.738673 | 32 |
| AACTC | 577040 | 1.2810911 | 13.932197 | 22 |
| ACGTC | 344355 | 1.2661833 | 22.475245 | 15 |
| CAGTC | 343910 | 1.264547 | 21.934828 | 27 |
| GAACT | 553840 | 1.2509955 | 14.186657 | 21 |
| ACAGT | 544745 | 1.2304521 | 13.822197 | 34 |
| TGATC | 544835 | 1.2297632 | 13.784545 | 38 |
| ATCTC | 546520 | 1.212454 | 13.618326 | 40 |
| GTGAT | 499020 | 1.145966 | 13.961749 | 37 |
| AGTCA | 490880 | 1.1087835 | 13.803366 | 28 |
| GATCT | 478495 | 1.0800252 | 13.725413 | 39 |
| TCGTA | 388290 | 0.8764209 | 13.591022 | 43 |
| TATGC | 371280 | 0.83802706 | 13.506125 | 46 |
| GTATG | 359100 | 0.8246492 | 13.727067 | 45 |
| CGTAT | 344470 | 0.77751344 | 13.465949 | 44 |