![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW233_TGACCA_L004_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 9726786 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 37 |
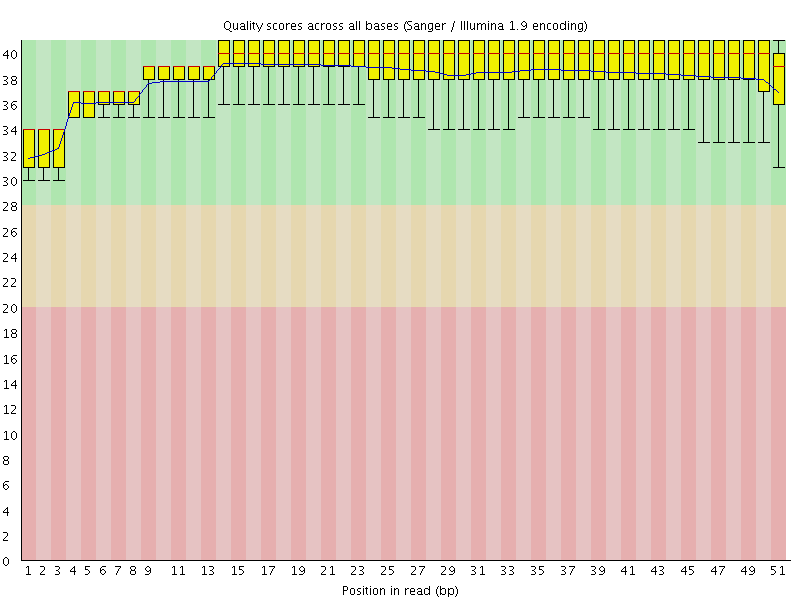
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
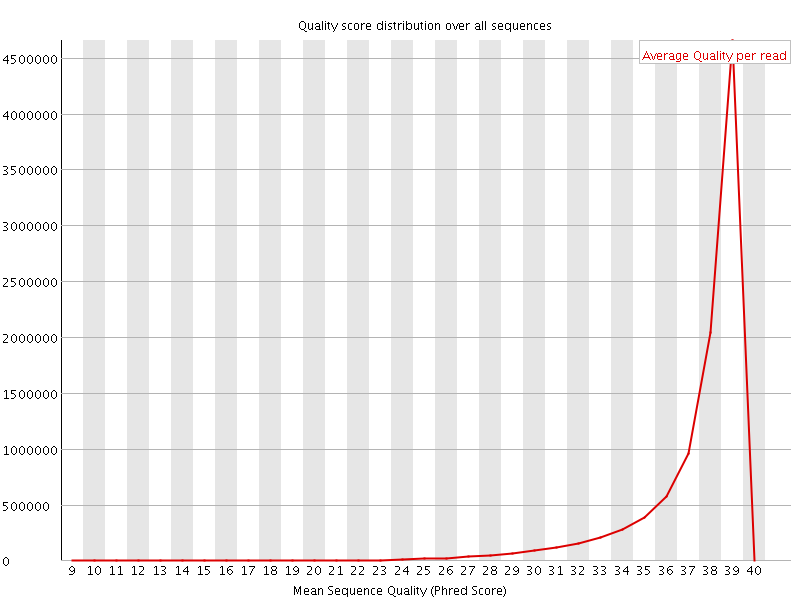
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
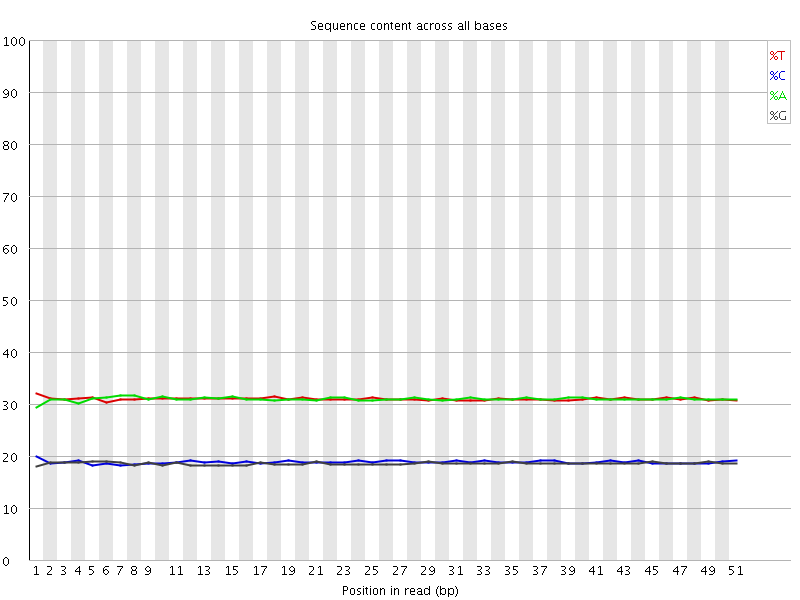
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
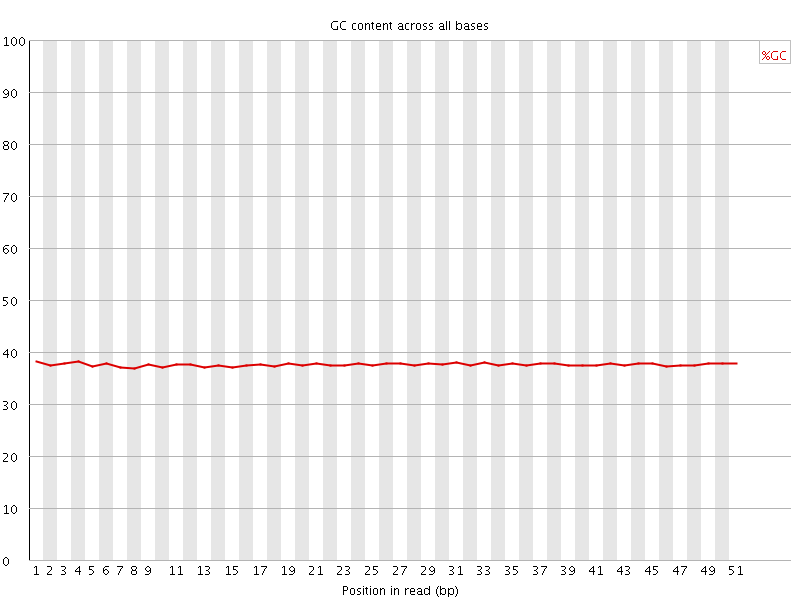
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
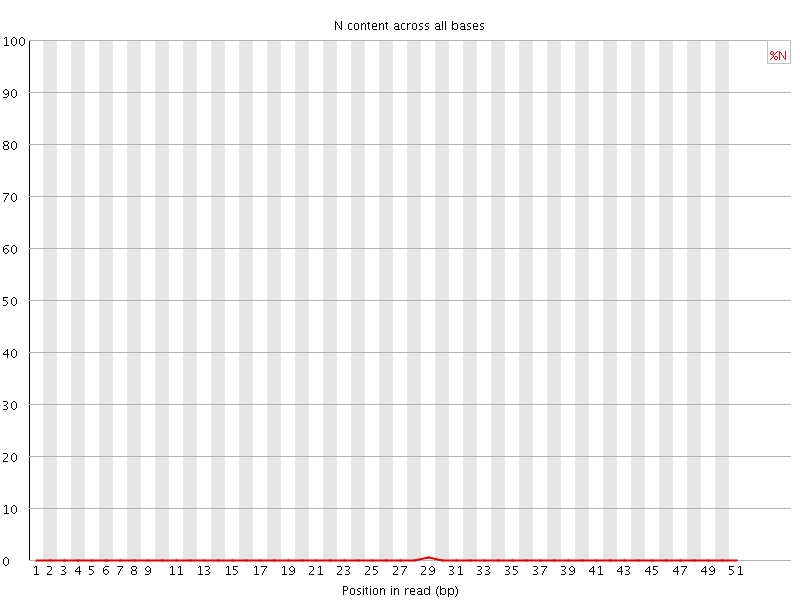
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
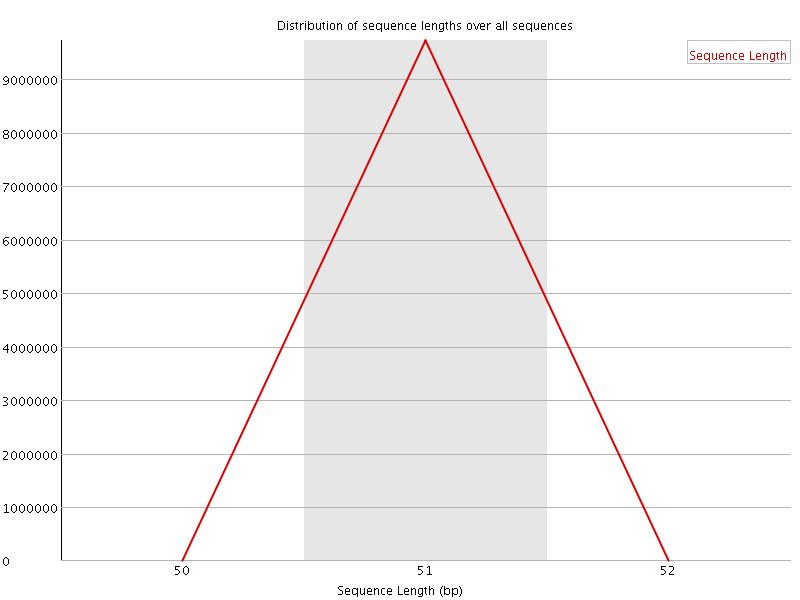
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACTGACCAATCTCGTATGCC | 35194 | 0.36182558144077603 | TruSeq Adapter, Index 4 (100% over 51bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
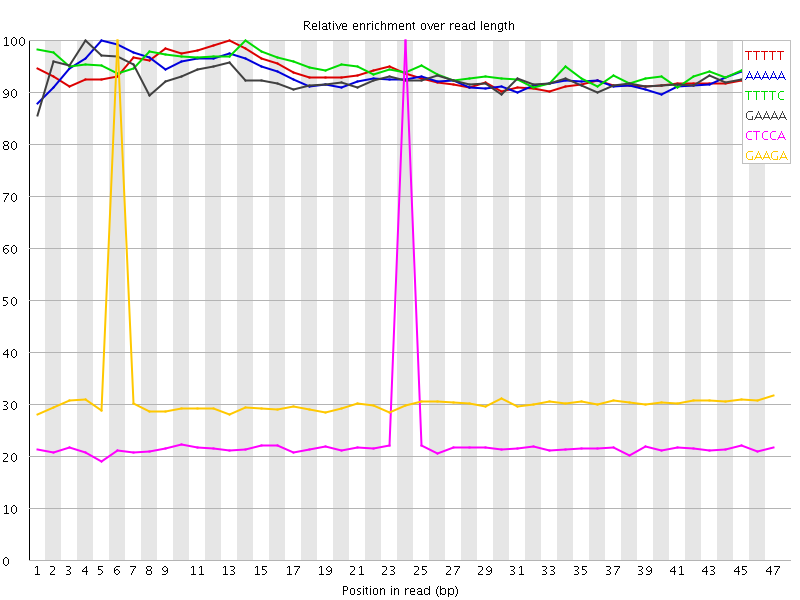
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 4721790 | 3.5242462 | 3.7702875 | 13 |
| AAAAA | 4686330 | 3.521779 | 3.7788656 | 5 |
| TTTTC | 2474735 | 3.0274403 | 3.2046943 | 14 |
| GAAAA | 2411755 | 3.009679 | 3.2509239 | 4 |
| CTCCA | 574040 | 1.8891118 | 8.152238 | 24 |
| GAAGA | 902695 | 1.8706211 | 5.946212 | 6 |
| CTGAA | 911460 | 1.8591911 | 5.8151097 | 19 |
| GAGCA | 543660 | 1.8440197 | 8.492828 | 9 |
| CGGAA | 532180 | 1.8050809 | 8.551756 | 4 |
| GGAAG | 517335 | 1.7802227 | 8.568552 | 5 |
| TCCAG | 522820 | 1.745549 | 8.123186 | 25 |
| TCTCG | 436375 | 1.4549423 | 7.65006 | 41 |
| TCGGA | 429260 | 1.4540008 | 8.10746 | 3 |
| TCTGA | 707460 | 1.441101 | 5.3870726 | 18 |
| GCACA | 401235 | 1.341444 | 7.8578653 | 11 |
| CACAC | 401960 | 1.3246229 | 7.7275405 | 12 |
| CACTG | 395830 | 1.3215652 | 7.5631275 | 31 |
| AGAGC | 384815 | 1.3052393 | 7.977605 | 8 |
| TGAAC | 614400 | 1.2532499 | 5.2039127 | 20 |
| AGCAC | 374135 | 1.2508411 | 7.829608 | 10 |
| ATCGG | 356520 | 1.207614 | 7.7575545 | 2 |
| AAGAG | 581345 | 1.2046995 | 5.2393026 | 7 |
| CCAGT | 356260 | 1.1894519 | 7.4960194 | 26 |
| GTCAC | 340715 | 1.1375517 | 7.393484 | 29 |
| CTCGT | 337495 | 1.125261 | 7.210599 | 42 |
| CACGT | 325705 | 1.0874374 | 7.5469117 | 14 |
| ACTCC | 327470 | 1.0776731 | 7.3807974 | 23 |
| CGTCT | 322280 | 1.0745318 | 7.532723 | 16 |
| GTCTG | 315840 | 1.0683595 | 7.667782 | 17 |
| CTGAC | 319790 | 1.0676888 | 7.176464 | 33 |
| GAACT | 522000 | 1.064773 | 5.0175977 | 21 |
| GACCA | 317320 | 1.0608922 | 7.1332383 | 35 |
| ACACG | 301090 | 1.0066307 | 7.496801 | 13 |
| ATGCC | 301035 | 1.0050712 | 7.1051917 | 47 |
| GATCG | 290920 | 0.98541176 | 7.4242883 | 1 |
| ACGTC | 290260 | 0.9690965 | 7.467781 | 15 |
| CAGTC | 288960 | 0.9647562 | 7.279075 | 27 |
| TGACC | 280730 | 0.9372785 | 6.9457836 | 34 |