![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW202_GTTTCG_L003_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 8528916 |
| Filtered Sequences | 0 |
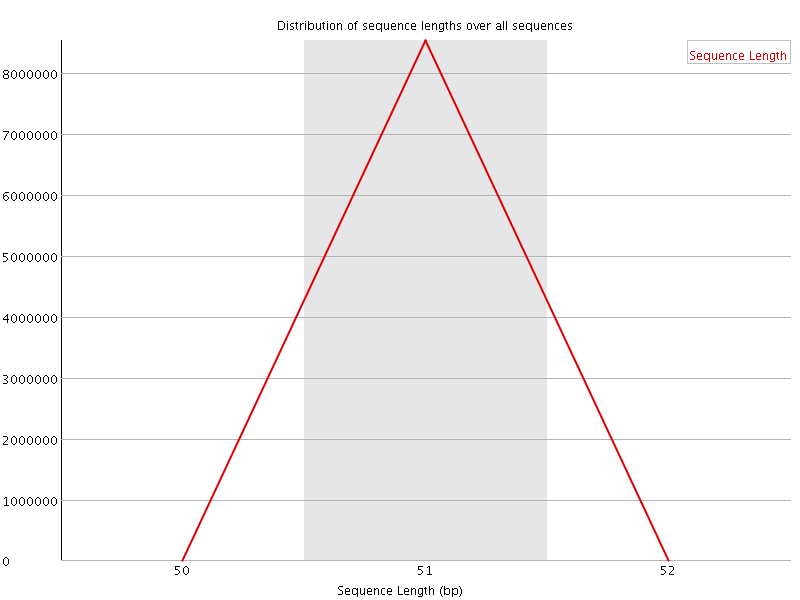
| Sequence length | 51 |
| %GC | 41 |
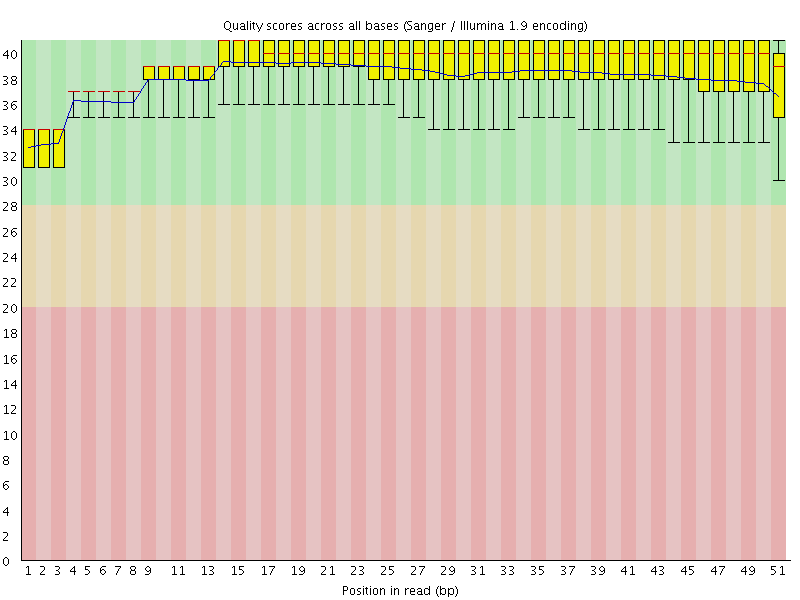
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
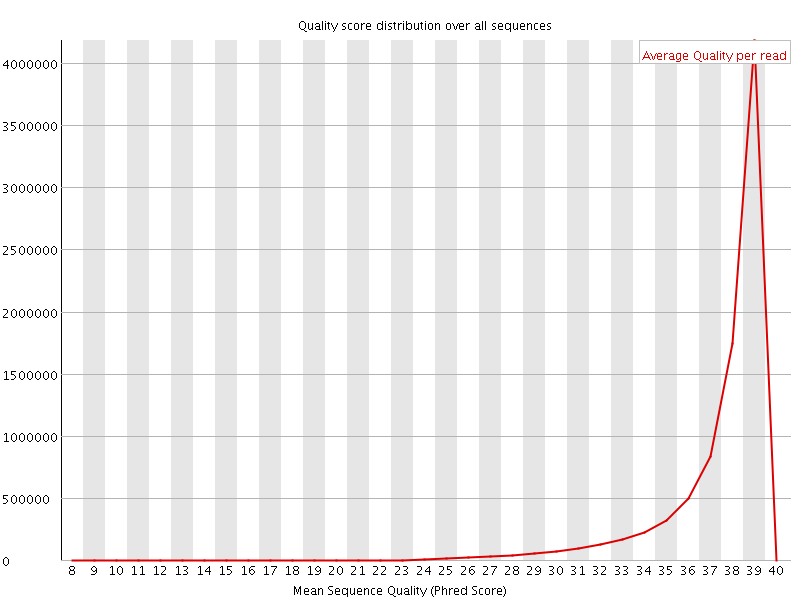
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
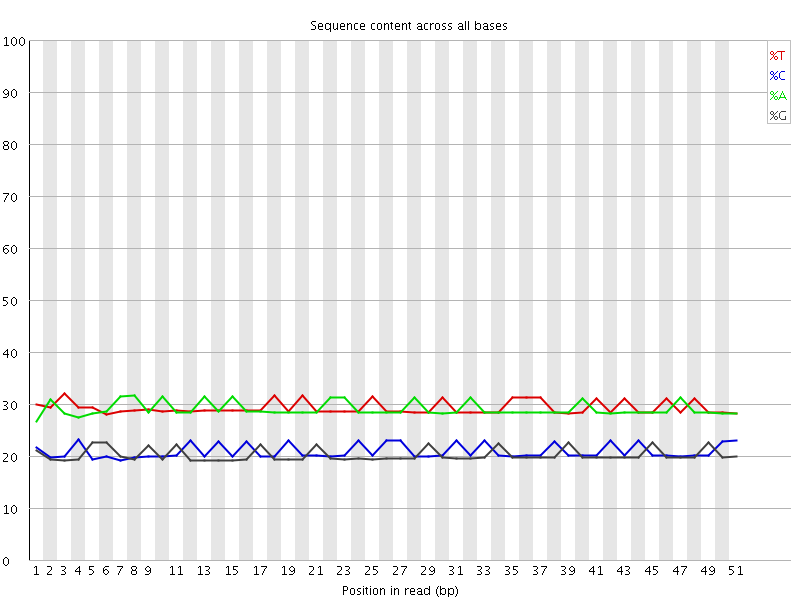
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
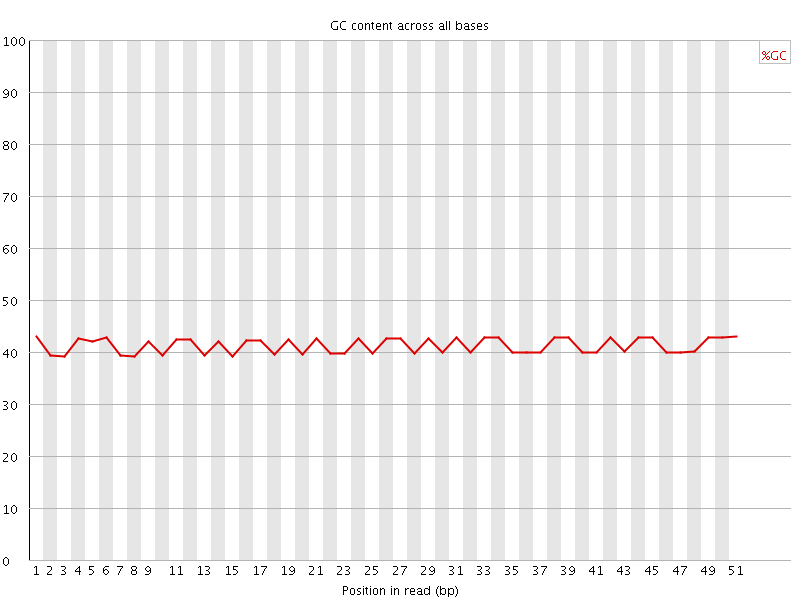
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
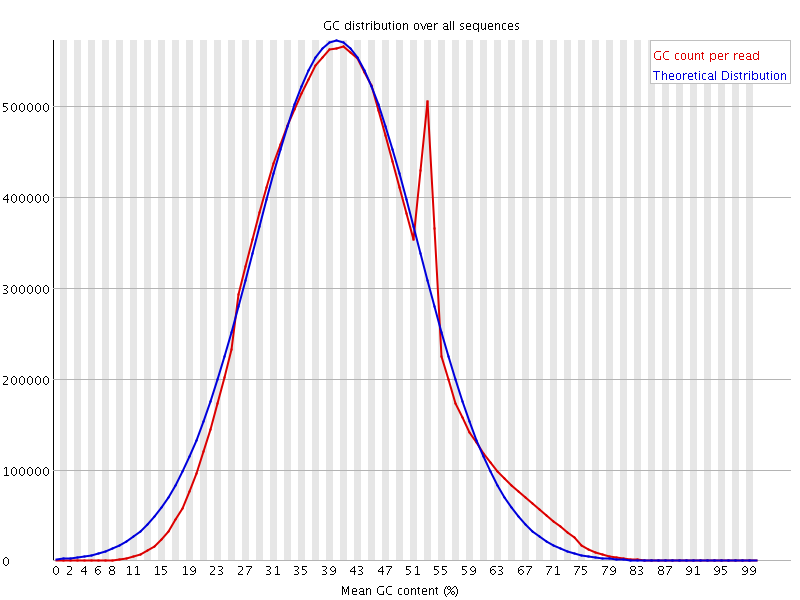
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
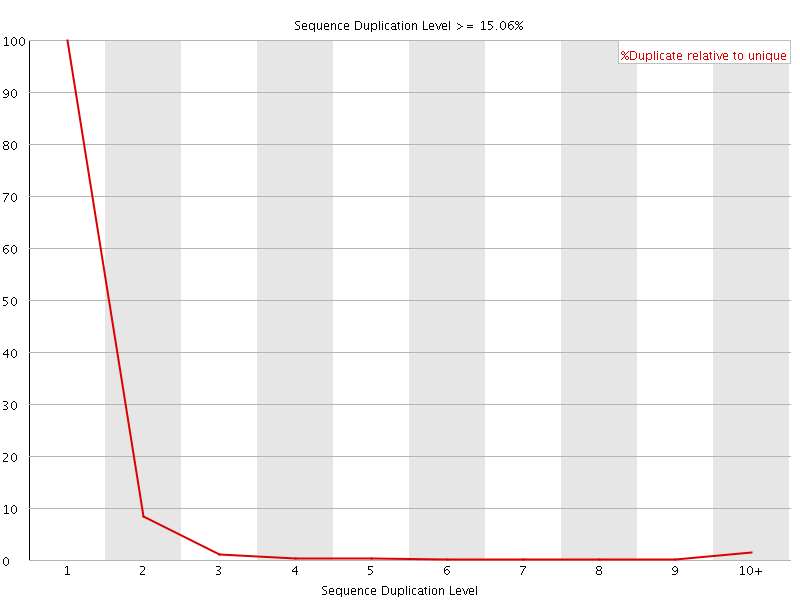
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACGTTTCGATCTCGTATGCC | 222887 | 2.613309827415348 | TruSeq Adapter, Index 9 (97% over 36bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 3079150 | 3.6362925 | 3.8350902 | 14 |
| TTTTT | 3136875 | 3.54688 | 3.870868 | 1 |
| CACGT | 767240 | 2.4767337 | 38.54087 | 14 |
| GAAGA | 1000635 | 2.428508 | 30.445347 | 6 |
| GAGCA | 709855 | 2.38931 | 41.16677 | 9 |
| GGAAG | 677120 | 2.3558493 | 42.64405 | 5 |
| CTCCA | 729470 | 2.2781196 | 37.53834 | 24 |
| CGGAA | 662180 | 2.2288399 | 41.244995 | 4 |
| TCCAG | 682990 | 2.2047656 | 38.68574 | 25 |
| CTGAA | 921460 | 2.1447942 | 28.619593 | 19 |
| TTTCG | 908090 | 2.0772395 | 27.187466 | 35 |
| TCTCG | 641395 | 2.0525696 | 37.746735 | 41 |
| TCGGA | 589300 | 1.9663625 | 40.667847 | 3 |
| AGAGC | 575645 | 1.9375707 | 40.75824 | 8 |
| CACAC | 611390 | 1.9260302 | 37.98773 | 12 |
| AGCAC | 589525 | 1.9196676 | 39.381947 | 10 |
| GATCG | 572465 | 1.9101876 | 40.260456 | 1 |
| TTCGA | 819490 | 1.890937 | 26.98858 | 36 |
| GTTTC | 823355 | 1.8834097 | 26.993305 | 34 |
| GCACA | 576780 | 1.878166 | 39.271095 | 11 |
| ATCGG | 560905 | 1.8716145 | 40.44851 | 2 |
| CGATC | 560020 | 1.8078051 | 36.91405 | 38 |
| AAGAG | 730775 | 1.7735668 | 29.719467 | 7 |
| TCTGA | 765800 | 1.7670497 | 28.05709 | 18 |
| CTCGT | 539365 | 1.7260568 | 37.25508 | 42 |
| CGTCT | 536730 | 1.7176243 | 38.215736 | 16 |
| CCAGT | 531955 | 1.7172083 | 38.04011 | 26 |
| TCGAT | 733025 | 1.6914228 | 26.827105 | 37 |
| GTCTG | 508605 | 1.6824107 | 39.34186 | 17 |
| ATGCC | 518335 | 1.6732414 | 37.40002 | 47 |
| GTCAC | 513380 | 1.6572461 | 37.97219 | 29 |
| TGAAC | 703640 | 1.6377953 | 28.162592 | 20 |
| ACGTC | 506565 | 1.6352466 | 38.46754 | 15 |
| CGTTT | 712085 | 1.6288815 | 26.990149 | 33 |
| ACACG | 497780 | 1.6209189 | 38.81778 | 13 |
| TCACG | 501685 | 1.6194936 | 37.7857 | 30 |
| ACTCC | 504240 | 1.574731 | 37.00371 | 23 |
| CAGTC | 480215 | 1.5501859 | 37.798195 | 27 |
| ATCTC | 680075 | 1.5181377 | 26.15598 | 40 |
| AACTC | 664345 | 1.4959732 | 27.080439 | 22 |
| GAACT | 641105 | 1.4922388 | 27.960363 | 21 |
| GATCT | 614940 | 1.4189469 | 26.817175 | 39 |
| AGTCA | 564560 | 1.3140721 | 27.428549 | 28 |
| ACGTT | 550395 | 1.2700121 | 26.891773 | 32 |
| TCGTA | 531685 | 1.2268398 | 26.770666 | 43 |
| TATGC | 488000 | 1.1260384 | 26.596786 | 46 |
| GTATG | 470720 | 1.1227293 | 27.508774 | 45 |
| CGTAT | 483430 | 1.1154934 | 26.649801 | 44 |