![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW195_AGTTCC_L002_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 7034302 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 43 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
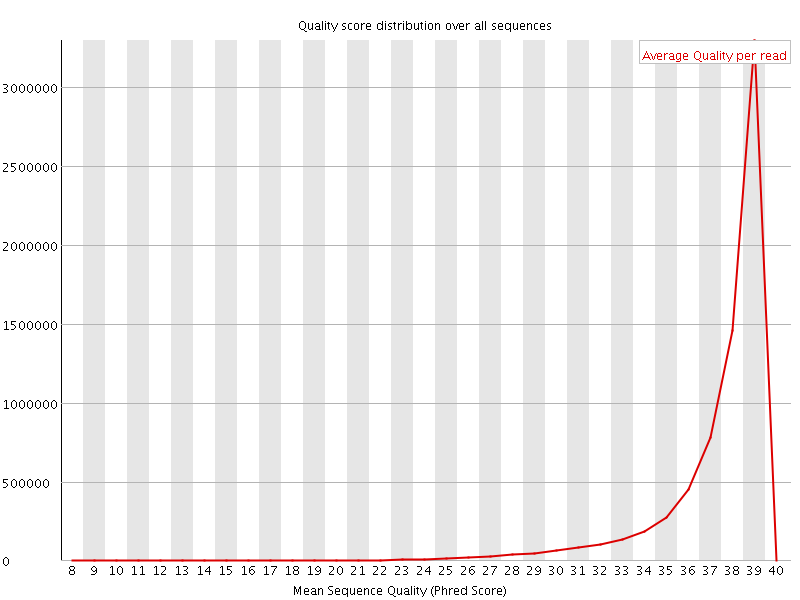
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
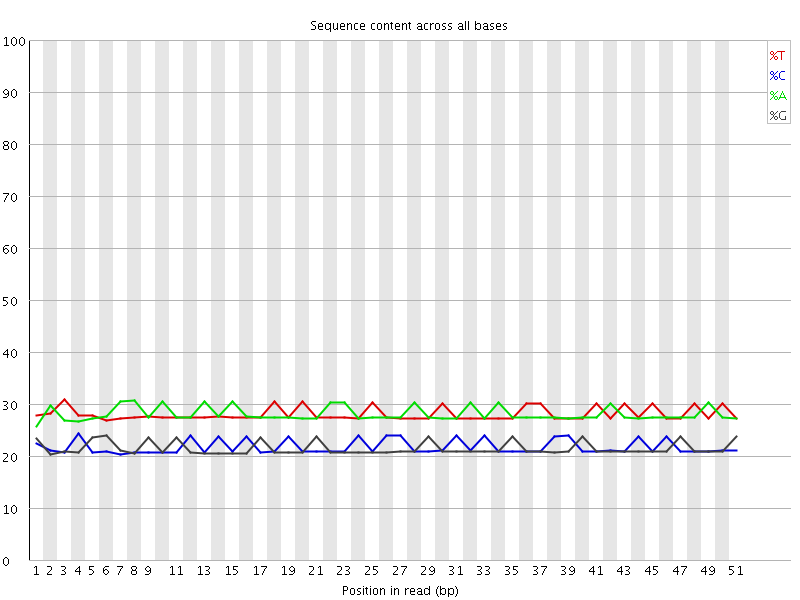
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
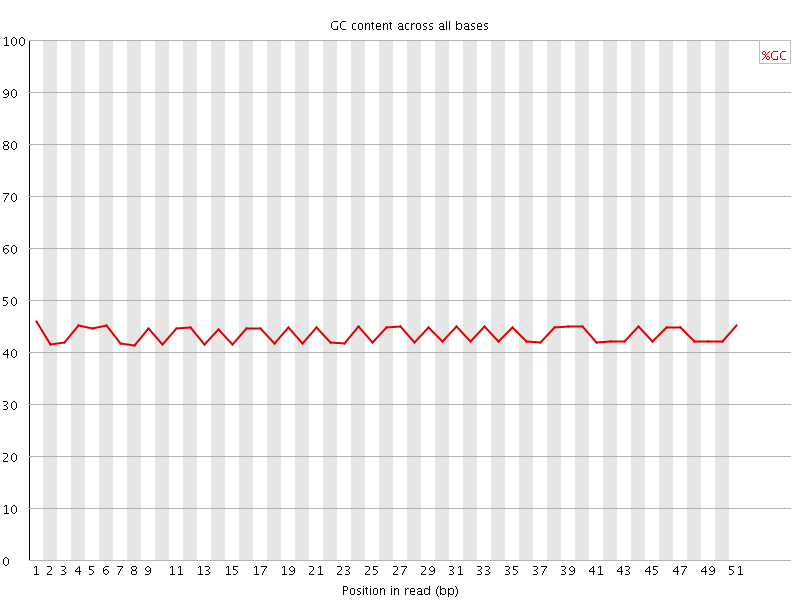
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
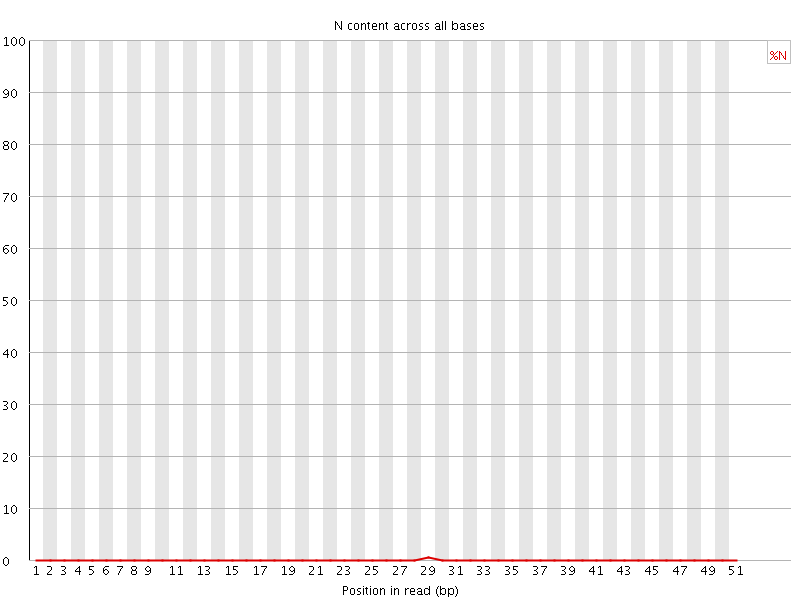
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
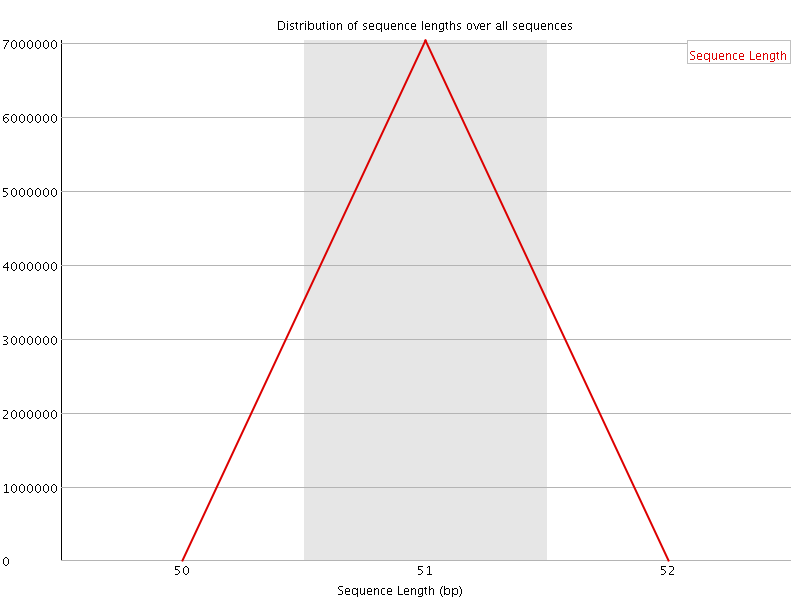
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACAGTTCCGTATCTCGTATG | 186089 | 2.645450820849034 | TruSeq Adapter, Index 8 (97% over 37bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
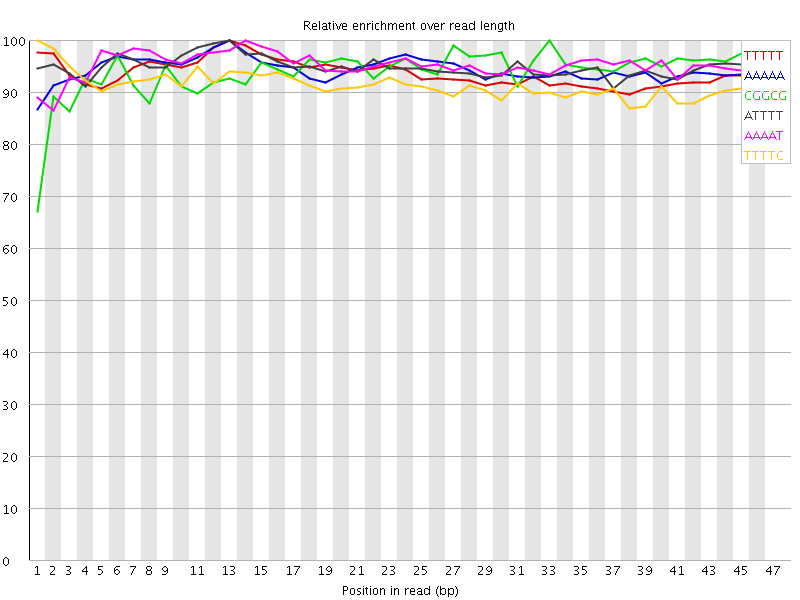
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 2799800 | 4.726517 | 5.041993 | 13 |
| AAAAA | 2802985 | 4.7068663 | 4.983001 | 13 |
| CGGCG | 549515 | 3.4386787 | 3.659612 | 33 |
| ATTTT | 2015070 | 3.398159 | 3.58064 | 13 |
| AAAAT | 2015155 | 3.3875067 | 3.5519023 | 14 |
| TTTTC | 1452760 | 3.1628163 | 3.4568536 | 1 |
| GAAAA | 1438015 | 3.1576004 | 3.28346 | 4 |
| CGCCG | 500620 | 3.0928876 | 3.4287088 | 45 |
| GATCG | 612245 | 2.2718976 | 38.378757 | 1 |
| TCCAG | 592400 | 2.1703131 | 37.09132 | 25 |
| CTGAA | 759050 | 2.1540277 | 29.455929 | 19 |
| CGGAA | 576360 | 2.1364696 | 38.38818 | 4 |
| GAAGA | 741635 | 2.1294458 | 30.152433 | 6 |
| GGAAG | 558915 | 2.09848 | 38.76171 | 5 |
| TTCCG | 569005 | 2.0868156 | 36.1013 | 36 |
| GAGCA | 555310 | 2.058441 | 38.07405 | 9 |
| TCTCG | 540060 | 1.9806603 | 36.27988 | 43 |
| ATCGG | 524420 | 1.9459996 | 38.178047 | 2 |
| CTCCA | 527360 | 1.9074728 | 36.413197 | 24 |
| TCGGA | 478135 | 1.774247 | 38.06394 | 3 |
| AGCAC | 473095 | 1.7313908 | 37.243835 | 10 |
| TCTGA | 606490 | 1.7229201 | 29.06233 | 18 |
| GTTCC | 462100 | 1.6947435 | 35.80441 | 35 |
| CAGTT | 587940 | 1.6702231 | 28.276527 | 33 |
| CCAGT | 455155 | 1.6675031 | 36.47902 | 26 |
| GCACA | 455290 | 1.6662297 | 37.14246 | 11 |
| AGAGC | 448935 | 1.6641265 | 37.615475 | 8 |
| CACAG | 449010 | 1.6432467 | 36.078094 | 31 |
| CGTAT | 572135 | 1.6253241 | 27.501114 | 46 |
| TGAAC | 572440 | 1.6244669 | 28.97119 | 20 |
| CACAC | 445845 | 1.6109219 | 36.547318 | 12 |
| GTCTG | 430285 | 1.5983813 | 37.3258 | 17 |
| GTCAC | 434915 | 1.5933522 | 36.277233 | 29 |
| CGTCT | 427650 | 1.5683987 | 36.80368 | 16 |
| ACGTC | 426275 | 1.5616986 | 36.88507 | 15 |
| CTCGT | 422945 | 1.5511433 | 35.742393 | 44 |
| AAGAG | 535160 | 1.5365971 | 29.421118 | 7 |
| ATCTC | 539070 | 1.5119258 | 27.589485 | 42 |
| GAACT | 528035 | 1.4984547 | 28.773312 | 21 |
| TCACA | 531755 | 1.4898285 | 27.9567 | 30 |
| AGTTC | 520370 | 1.4782699 | 27.98177 | 34 |
| CACGT | 403330 | 1.4776374 | 36.85163 | 14 |
| TCCGT | 402340 | 1.4755749 | 35.551605 | 37 |
| ACACG | 393980 | 1.4418529 | 36.8581 | 13 |
| AACTC | 511085 | 1.431917 | 28.376501 | 22 |
| CAGTC | 385590 | 1.4126453 | 36.214874 | 27 |
| ACTCC | 387875 | 1.4029527 | 36.078526 | 23 |
| ACAGT | 471640 | 1.3384173 | 27.956388 | 32 |
| CCGTA | 348330 | 1.2761396 | 35.152203 | 38 |
| AGTCA | 445050 | 1.2629603 | 28.161802 | 28 |
| TCGTA | 385425 | 1.0949174 | 27.511765 | 45 |
| GTATC | 379975 | 1.079435 | 27.452328 | 40 |
| GTATG | 364180 | 1.0478854 | 27.728018 | 47 |
| TATCT | 461735 | 1.0041816 | 21.204065 | 41 |