![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | LW179_CGTACG_L002_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 8391404 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 36 |
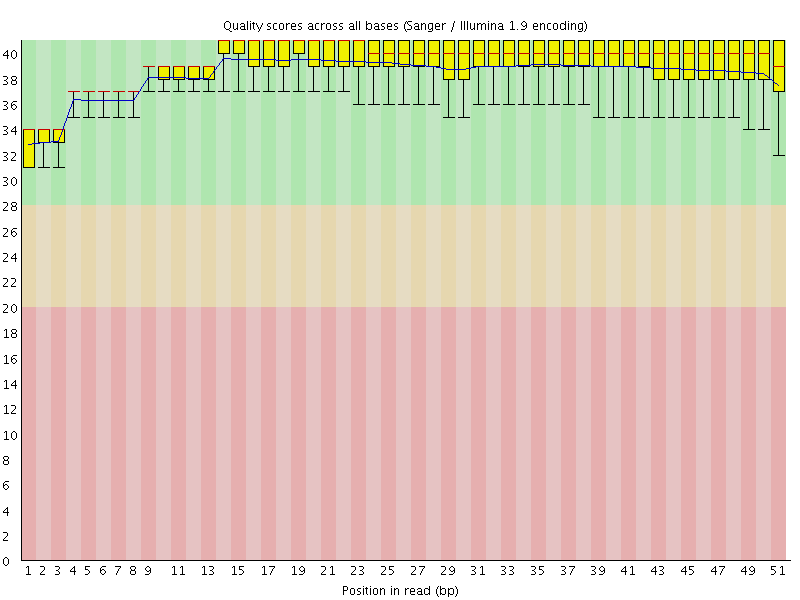
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
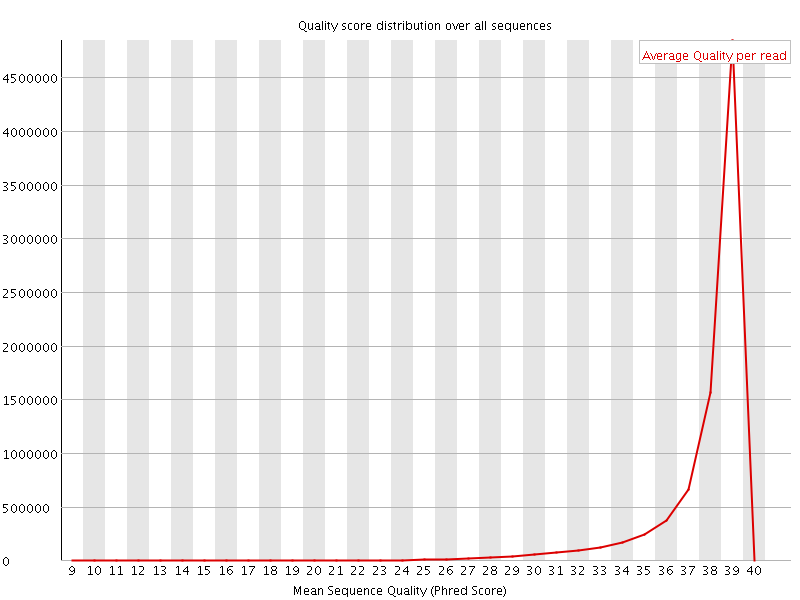
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
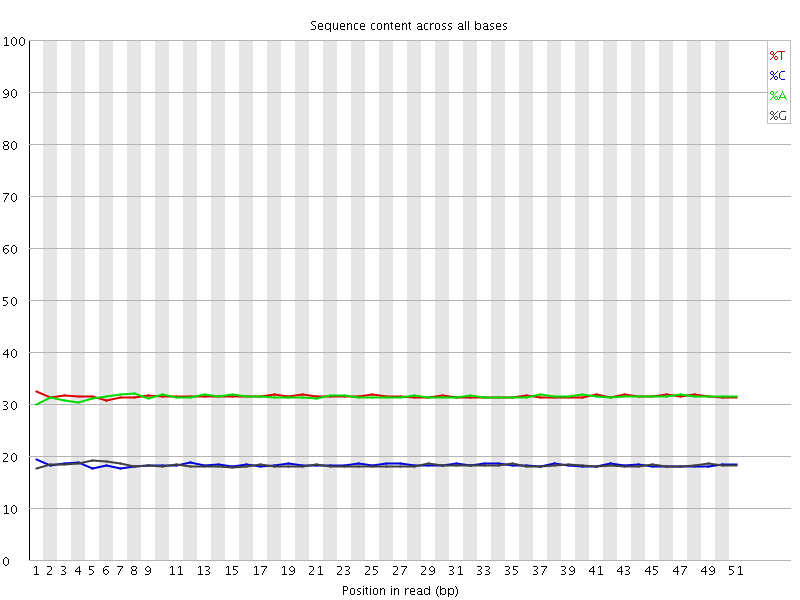
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
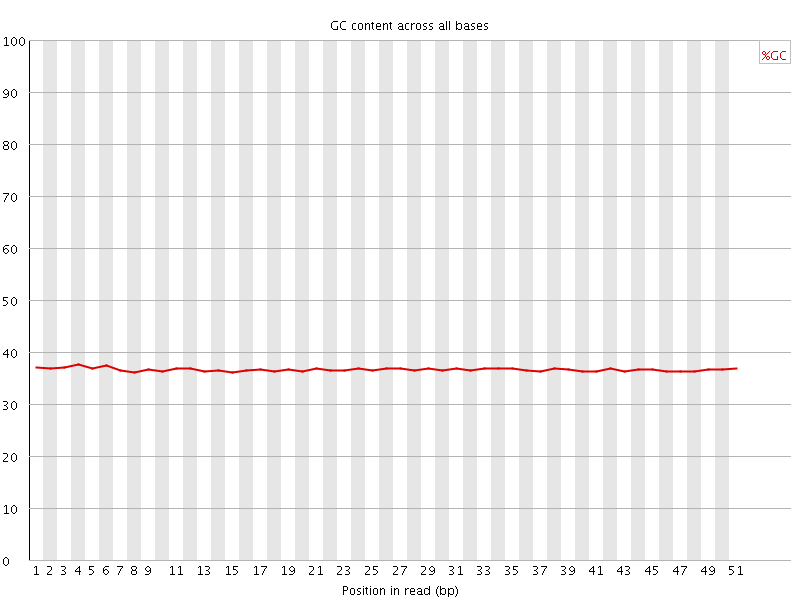
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
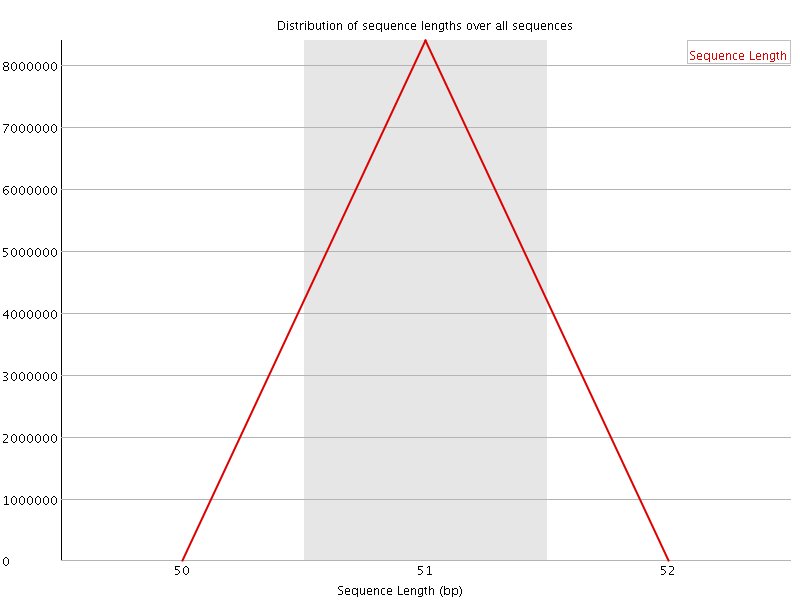
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACCGTACGATCTCGTATGCC | 30821 | 0.36729252935504 | TruSeq Adapter, Index 12 (97% over 36bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
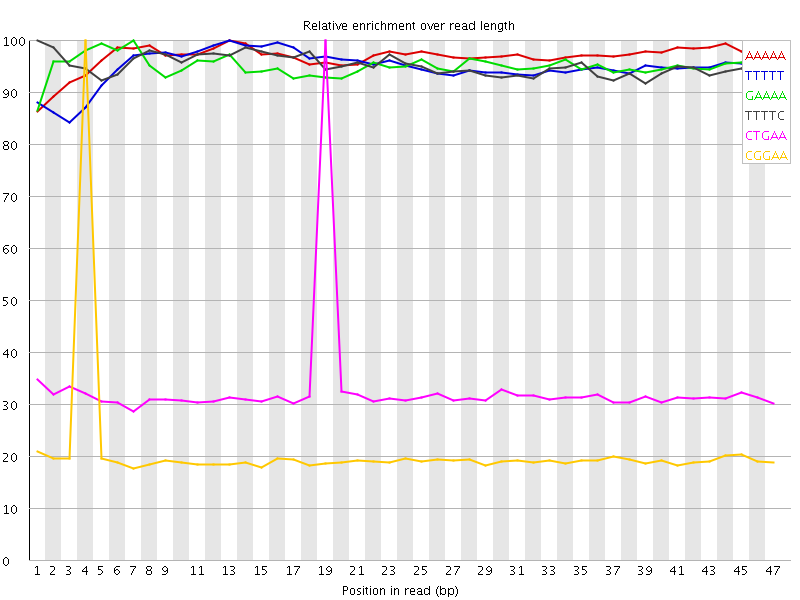
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 4510750 | 3.6754017 | 3.7926776 | 13 |
| TTTTT | 4605480 | 3.6738255 | 3.870367 | 13 |
| GAAAA | 2242575 | 3.1376705 | 3.2991297 | 7 |
| TTTTC | 2285470 | 3.1288948 | 3.2851758 | 1 |
| CTGAA | 807600 | 1.9228415 | 5.860462 | 19 |
| CGGAA | 458945 | 1.8843198 | 9.024942 | 4 |
| CTCCA | 460025 | 1.8628724 | 8.688654 | 24 |
| GAAGA | 764715 | 1.837233 | 5.9182696 | 6 |
| GGAAG | 438020 | 1.8070195 | 8.966481 | 5 |
| GAGCA | 439230 | 1.8033746 | 8.779176 | 9 |
| TCCAG | 433450 | 1.7636629 | 8.5383835 | 25 |
| TCACC | 366395 | 1.4837174 | 7.959529 | 30 |
| TCTCG | 364635 | 1.4773806 | 7.893705 | 41 |
| TCGGA | 360525 | 1.4739639 | 8.544308 | 3 |
| TCTGA | 618580 | 1.4665626 | 5.415957 | 18 |
| CACCG | 205340 | 1.4339104 | 12.44355 | 31 |
| GCACA | 330220 | 1.3493432 | 8.266367 | 11 |
| CACAC | 326260 | 1.3268076 | 8.24224 | 12 |
| AGAGC | 317155 | 1.3021636 | 8.256802 | 8 |
| TGAAC | 521730 | 1.2422043 | 5.1812825 | 20 |
| CCAGT | 297890 | 1.2120835 | 7.949662 | 26 |
| ATCGG | 294385 | 1.2035584 | 8.201693 | 2 |
| AGCAC | 293175 | 1.1979702 | 8.161814 | 10 |
| AAGAG | 490570 | 1.1785978 | 5.2076716 | 7 |
| GTCAC | 276895 | 1.1266569 | 7.6235523 | 29 |
| CTCGT | 272420 | 1.103756 | 7.4431753 | 42 |
| AACTC | 465680 | 1.1034684 | 5.0793433 | 22 |
| ACTCC | 269185 | 1.0900652 | 7.8599353 | 23 |
| GTCTG | 266590 | 1.0853077 | 7.930257 | 17 |
| CACGT | 262880 | 1.0696313 | 7.9018483 | 14 |
| GAACT | 448275 | 1.0673128 | 5.056068 | 21 |
| CGTCT | 262130 | 1.0620642 | 7.860875 | 16 |
| ATGCC | 248000 | 1.0090861 | 7.364435 | 47 |
| ACACG | 244875 | 1.0006069 | 7.928729 | 13 |
| CAGTC | 235685 | 0.95897776 | 7.63221 | 27 |
| GATCG | 232310 | 0.949772 | 7.8701615 | 1 |
| ACGTC | 233270 | 0.94915134 | 7.8245707 | 15 |
| CGATC | 226600 | 0.9220118 | 7.24226 | 38 |
| ACCGT | 219860 | 0.89458746 | 7.261074 | 32 |
| CCGTA | 192705 | 0.78409666 | 7.122354 | 33 |
| GTACG | 153860 | 0.6290385 | 7.069686 | 35 |
| CGTAC | 154085 | 0.6269558 | 6.9661984 | 34 |