![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 140701_JD_C_14_TGACCA_L002_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 20892507 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 46 |
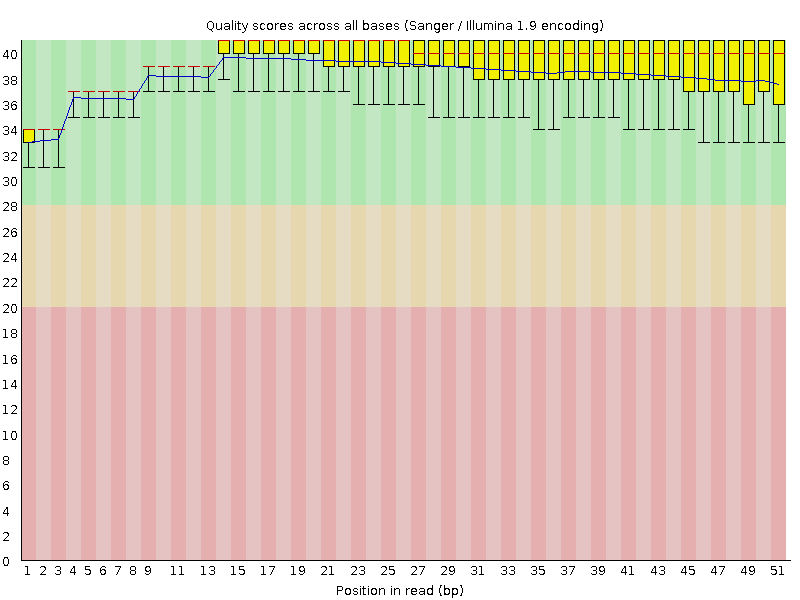
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
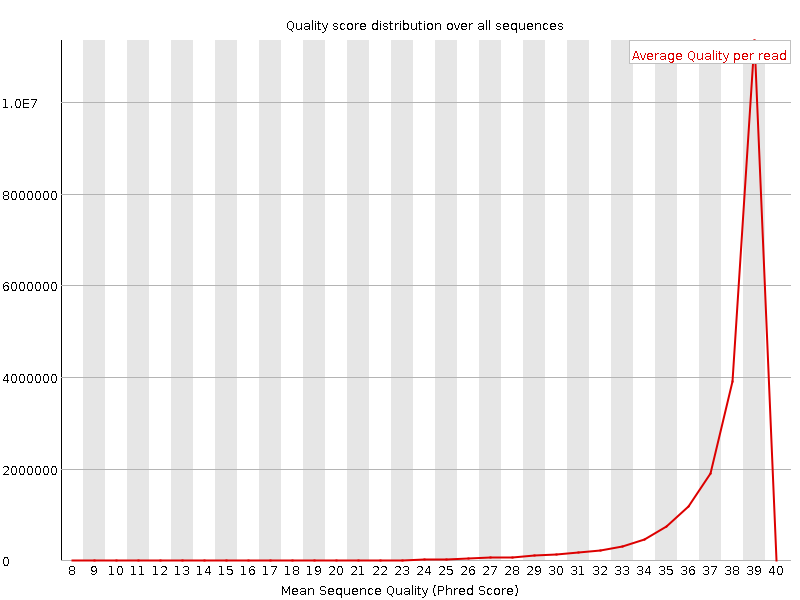
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[OK]](Icons/tick.png) Per base GC content
Per base GC content

![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution

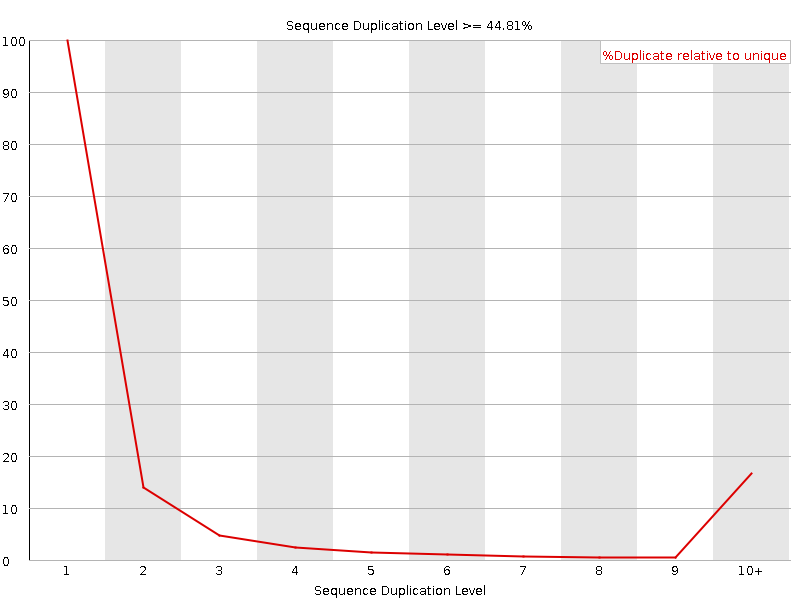
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTGTGTTTGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 34899 | 0.16704074814956385 | No Hit |
| CTTTGTGTTTGACGGGCGGTGTGTACAAAGGGCAGGGACTTAATCAACGCA | 24787 | 0.11864062077375395 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
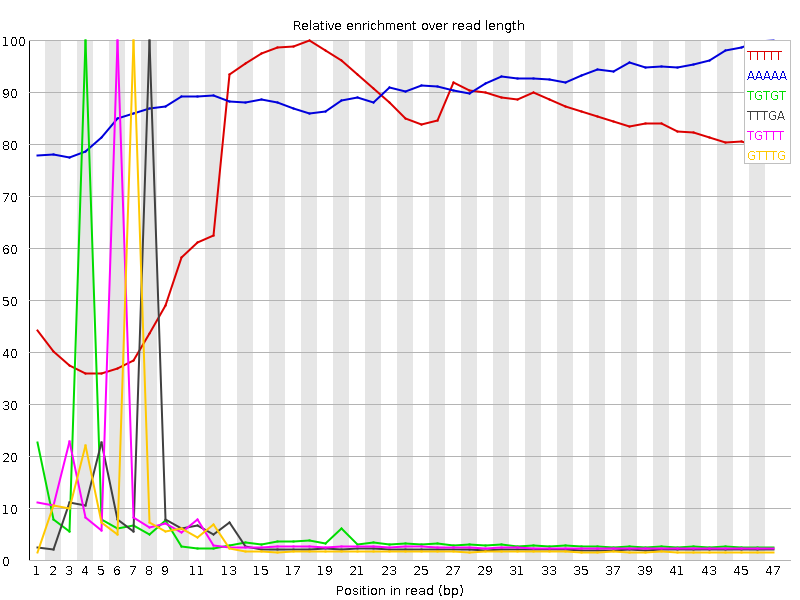
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 8504030 | 5.4878235 | 7.081896 | 18 |
| AAAAA | 4618945 | 4.103625 | 4.5515127 | 47 |
| TGTGT | 3984350 | 3.388609 | 55.550877 | 4 |
| TTTGA | 3782485 | 2.9872167 | 51.592567 | 8 |
| TGTTT | 3997605 | 2.9615533 | 48.539074 | 6 |
| GTTTG | 3450915 | 2.9349332 | 55.23611 | 7 |
| CTTTG | 3036210 | 2.727411 | 58.08661 | 1 |
| GTGTT | 3174730 | 2.7000437 | 55.16902 | 5 |
| TTTGT | 3252925 | 2.4098704 | 48.131348 | 2 |
| TTGTG | 2706460 | 2.301789 | 55.05566 | 3 |
| GAGGG | 1714250 | 2.0483139 | 12.030679 | 11 |
| TTGAG | 2241265 | 2.0320153 | 32.58116 | 9 |
| TGAGG | 1663985 | 1.7319188 | 17.497517 | 10 |
| TGAGC | 1266115 | 1.3918941 | 7.2946453 | 10 |
| AGGGG | 1120295 | 1.3386121 | 5.958892 | 12 |
| TTGAT | 1679235 | 1.3261755 | 11.699215 | 9 |
| TGAGA | 1330215 | 1.2856584 | 5.5994577 | 10 |
| TGAGT | 1248980 | 1.1323726 | 8.735266 | 10 |
| TGATT | 1406745 | 1.1109768 | 5.7527366 | 10 |
| TTGAC | 1159050 | 1.1099179 | 11.697569 | 9 |
| GAGTG | 1065420 | 1.1089168 | 5.6878786 | 11 |
| TGACG | 495970 | 0.5452409 | 5.4336553 | 10 |