![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 140701_JD_B_2_TGACCA_L008_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 34423689 |
| Filtered Sequences | 0 |
| Sequence length | 51 |
| %GC | 43 |
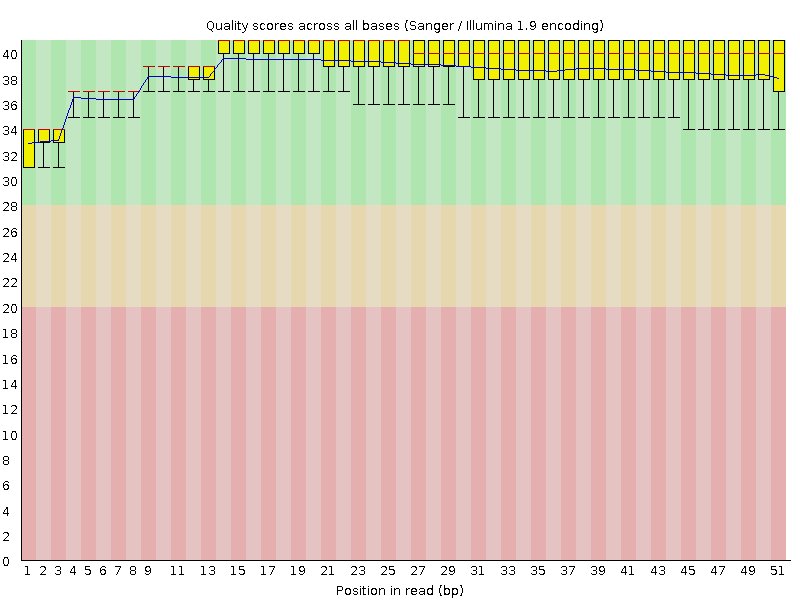
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
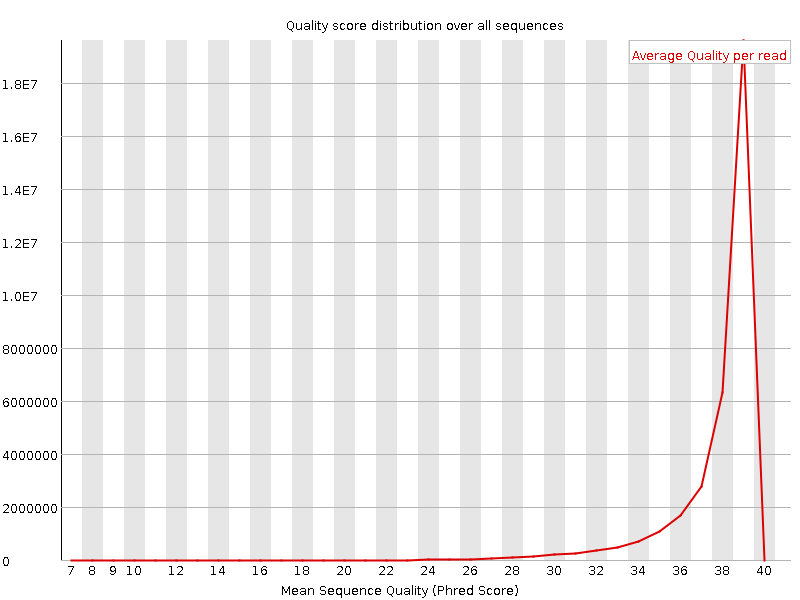
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content

![[OK]](Icons/tick.png) Per base GC content
Per base GC content

![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content

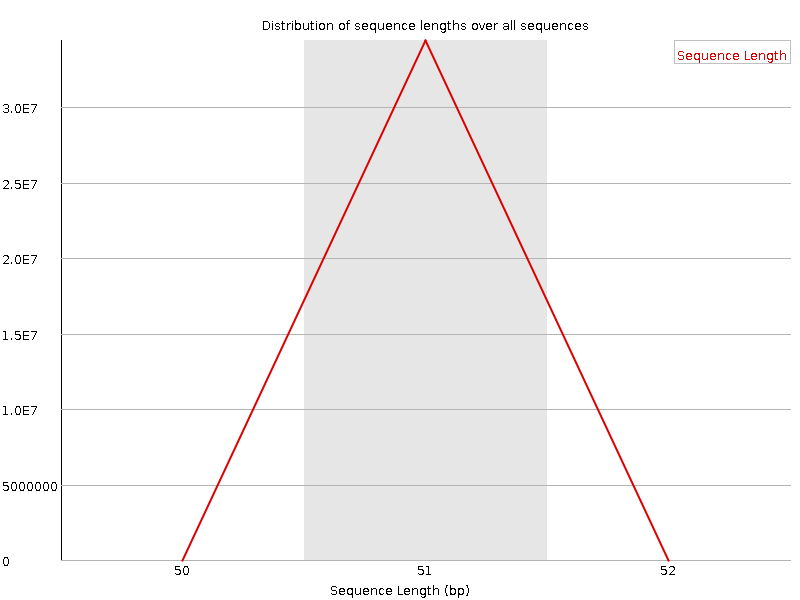
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
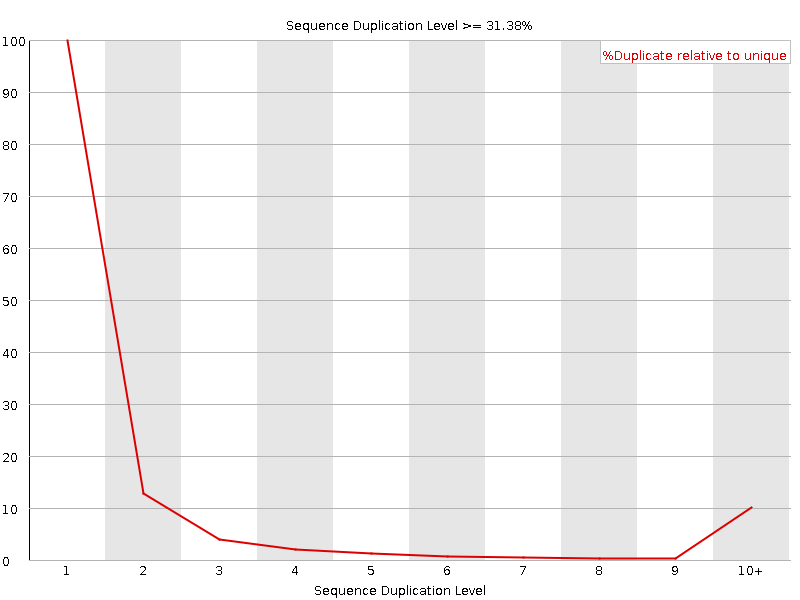
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTGTGTTTGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 51597 | 0.14988806109653152 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
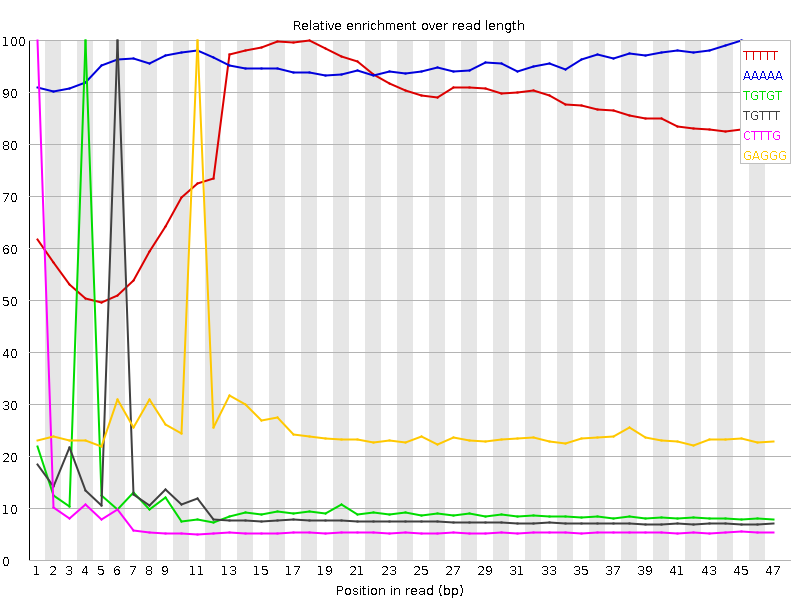
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 13341040 | 4.390675 | 5.3245187 | 18 |
| AAAAA | 8391425 | 3.177907 | 3.3259494 | 45 |
| TGTGT | 5352805 | 2.8910732 | 25.527302 | 4 |
| TGTTT | 5129305 | 2.16256 | 20.078823 | 6 |
| CTTTG | 3619095 | 2.0166278 | 25.687517 | 1 |
| GAGGG | 2176210 | 1.9838428 | 7.6361184 | 11 |
| GTTTG | 3609310 | 1.949404 | 24.833317 | 7 |
| TTTGT | 4317070 | 1.8201147 | 19.764532 | 2 |
| TTTGA | 4193965 | 1.8185582 | 20.199917 | 8 |
| GTGTT | 3292495 | 1.7782909 | 24.730665 | 5 |
| TGAGG | 2376925 | 1.6914294 | 9.3884945 | 10 |
| TTGTG | 3107970 | 1.6786281 | 24.772293 | 3 |
| TTGAG | 2692215 | 1.4954785 | 14.877455 | 9 |
| TTGAT | 2291230 | 0.9935074 | 5.3845863 | 9 |