![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | ZL1_CGATGT_L008_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 12906465 |
| Filtered Sequences | 0 |
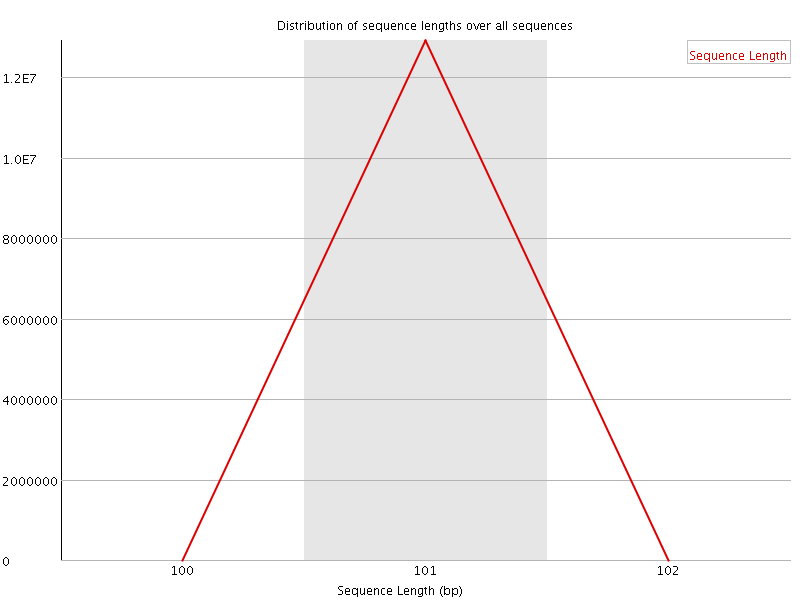
| Sequence length | 101 |
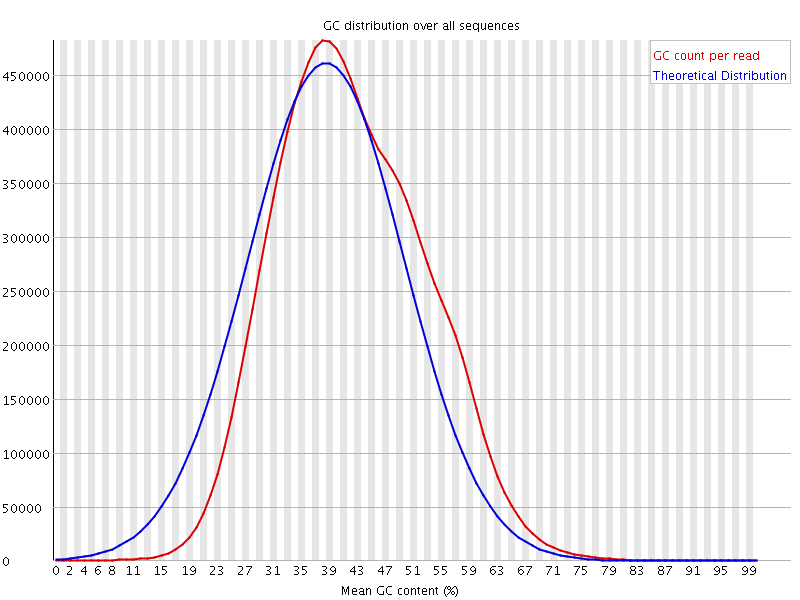
| %GC | 41 |
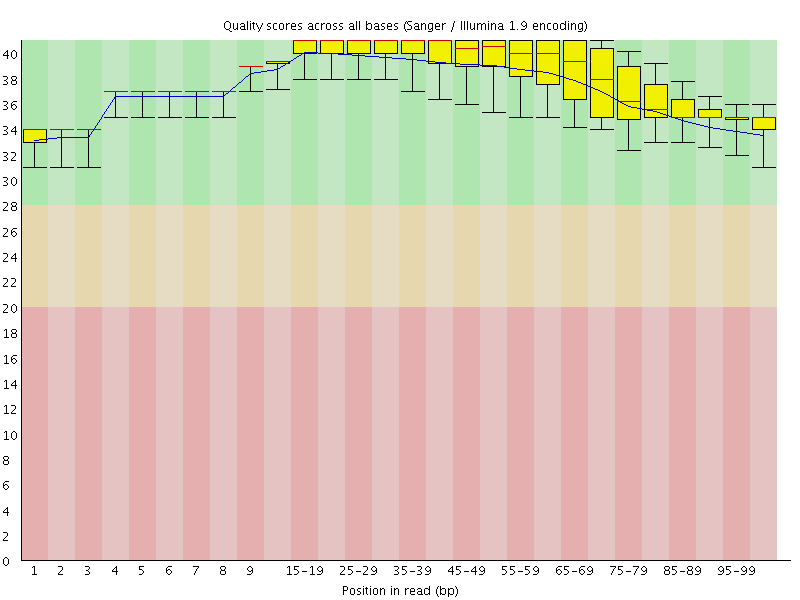
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
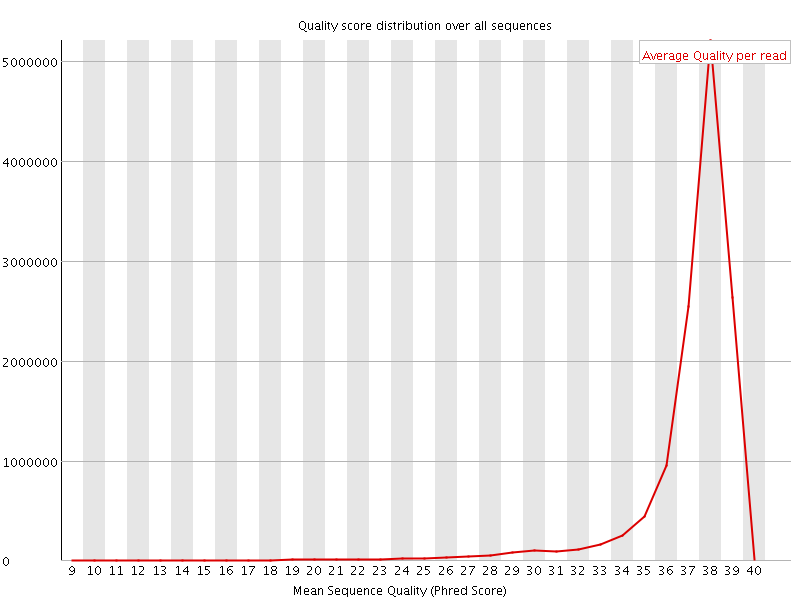
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
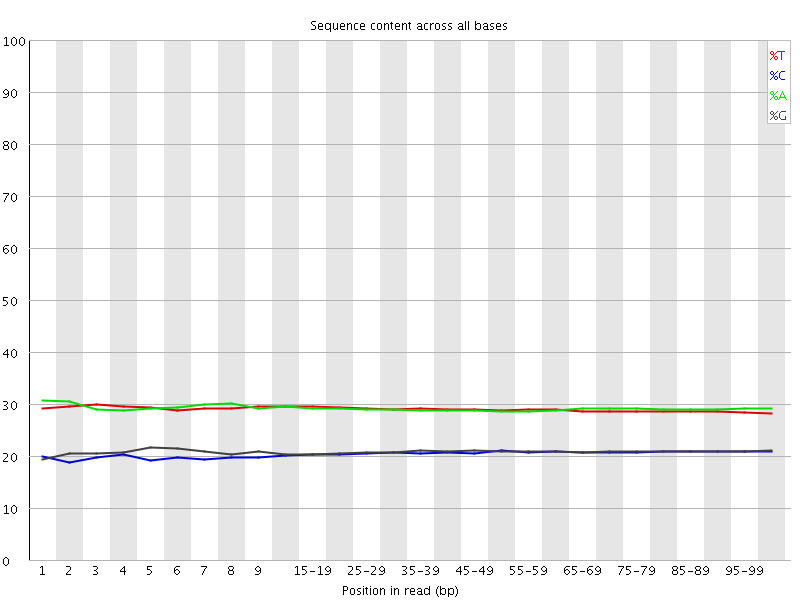
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
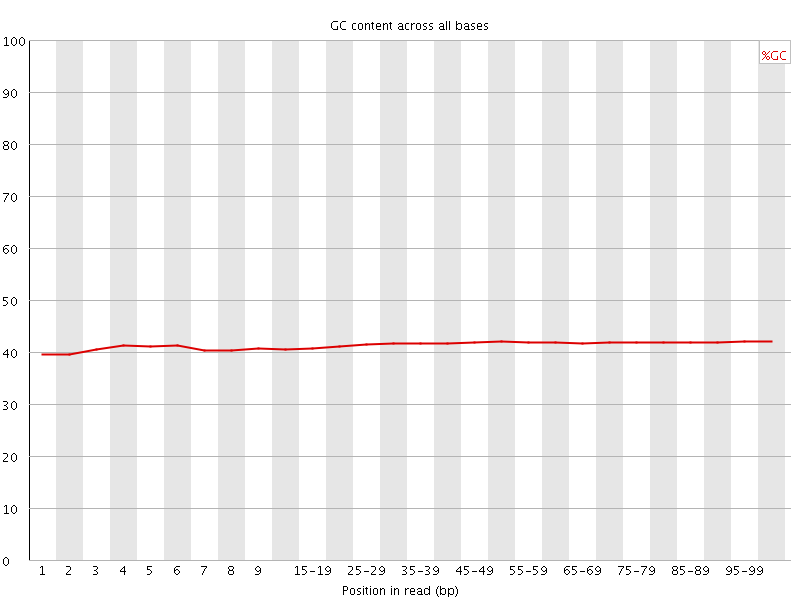
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
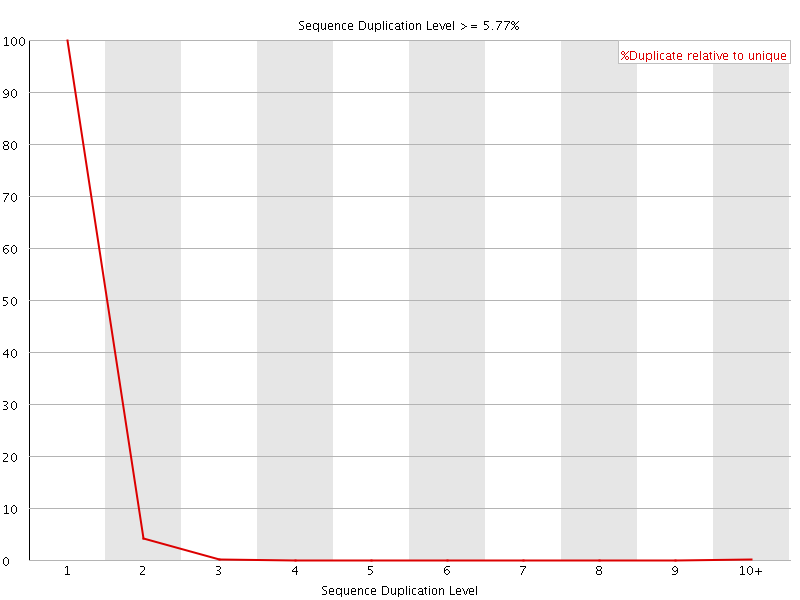
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACCGATGTATCTCGTATGC | 79581 | 0.6165979607894183 | TruSeq Adapter, Index 2 (100% over 50bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
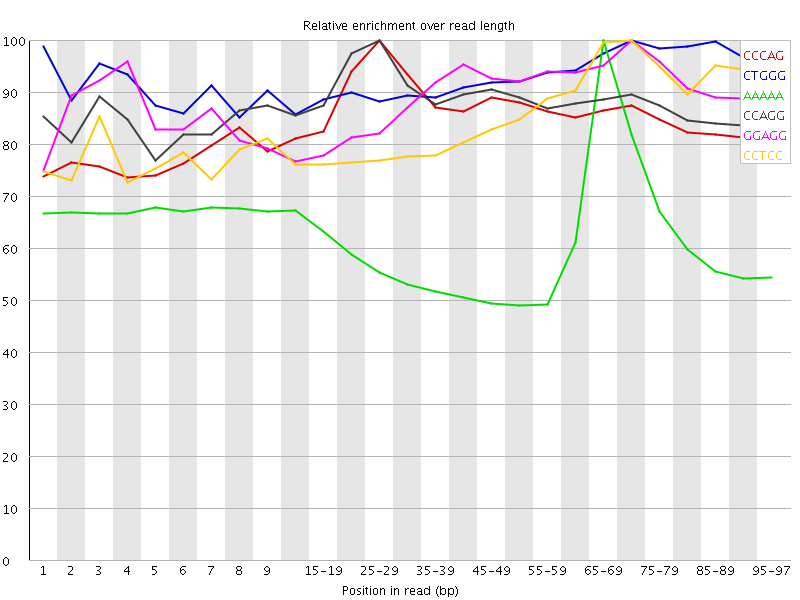
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCCAG | 2550695 | 3.724268 | 4.336115 | 25-29 |
| CTGGG | 2445860 | 3.518418 | 3.7749634 | 70-74 |
| AAAAA | 9119410 | 3.4300857 | 5.62669 | 65-69 |
| CCAGG | 2286485 | 3.3072739 | 3.7438 | 25-29 |
| GGAGG | 2320115 | 3.2934425 | 3.6874993 | 70-74 |
| CCTCC | 2195030 | 3.2478662 | 3.7963243 | 70-74 |
| CCTGG | 2154725 | 3.1288755 | 3.402336 | 70-74 |
| TTTTT | 8135325 | 3.1202292 | 3.7810225 | 10-14 |
| GGAAG | 2041300 | 2.0789633 | 10.501784 | 5 |
| GAAGA | 2137890 | 1.5621572 | 7.6352763 | 6 |
| AAGAG | 2132910 | 1.5585183 | 7.5604815 | 7 |
| AGAGC | 1418885 | 1.458706 | 9.828617 | 8 |
| GAGCA | 1370885 | 1.409359 | 9.797208 | 9 |
| GATCG | 433455 | 0.44736224 | 8.882546 | 1 |
| CGGAA | 404000 | 0.41533828 | 8.882384 | 4 |
| TCGGA | 361105 | 0.3726909 | 8.863615 | 3 |
| ATCGG | 352935 | 0.36425877 | 8.851104 | 2 |