![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | ZL1_CGATGT_L007_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 13280025 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 41 |
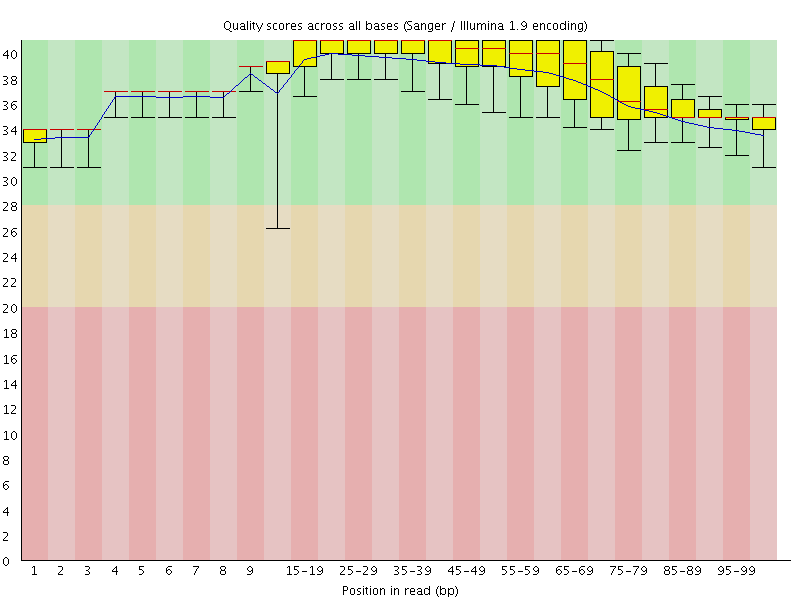
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
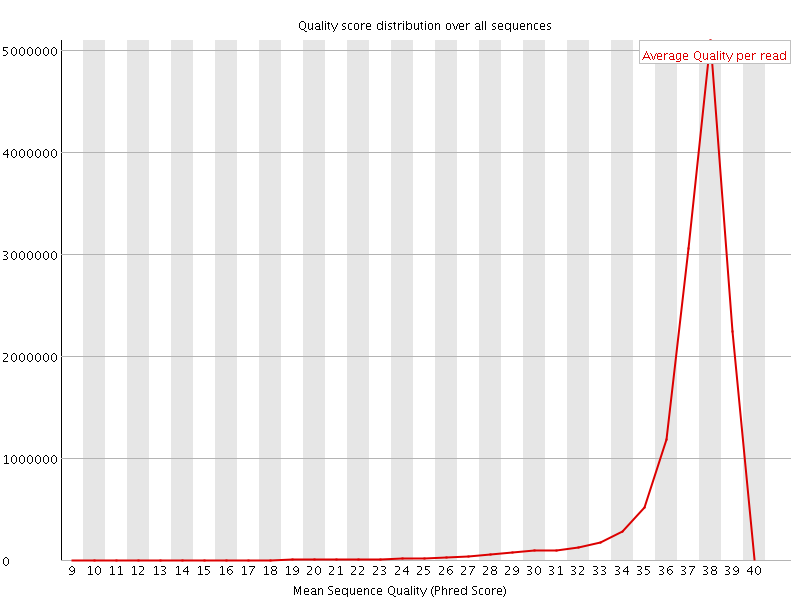
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
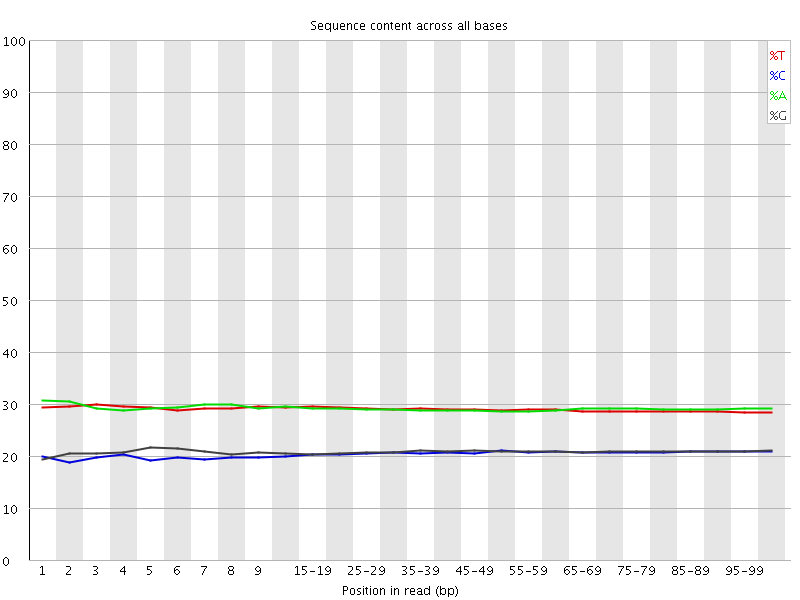
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
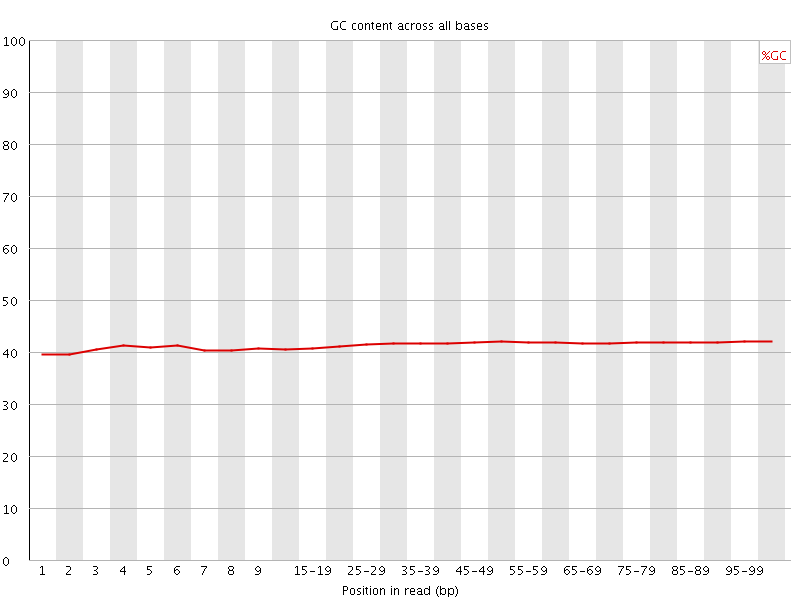
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACCGATGTATCTCGTATGC | 68529 | 0.5160306550627728 | TruSeq Adapter, Index 2 (100% over 50bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
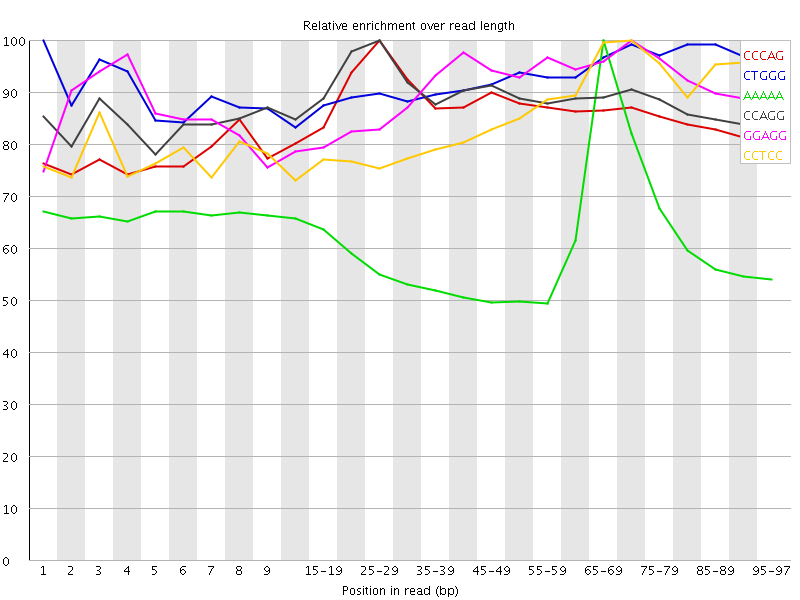
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCCAG | 2599050 | 3.7192237 | 4.312791 | 25-29 |
| CTGGG | 2497610 | 3.518772 | 3.7935221 | 1 |
| AAAAA | 9331440 | 3.4216545 | 5.6120477 | 65-69 |
| CCAGG | 2332880 | 3.3052142 | 3.720733 | 25-29 |
| GGAGG | 2373775 | 3.2967484 | 3.6454194 | 70-74 |
| CCTCC | 2237410 | 3.2479064 | 3.7974238 | 70-74 |
| CCTGG | 2198225 | 3.1280174 | 3.3967369 | 70-74 |
| TTTTT | 8270265 | 3.099245 | 3.7262764 | 15-19 |
| GGAAG | 2085585 | 2.076273 | 10.300786 | 5 |
| GAAGA | 2186905 | 1.5606188 | 7.436434 | 6 |
| AAGAG | 2180050 | 1.5557269 | 7.4123645 | 7 |
| AGAGC | 1443300 | 1.4512547 | 9.631072 | 8 |
| GAGCA | 1387355 | 1.3950014 | 9.399167 | 9 |
| GATCG | 441820 | 0.4461924 | 8.714833 | 1 |
| CGGAA | 411410 | 0.41367742 | 8.700136 | 4 |
| TCGGA | 367540 | 0.37117732 | 8.68259 | 3 |
| ATCGG | 358260 | 0.3618055 | 8.67091 | 2 |