![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-183_AAGAGGCA-AAGGAGTA_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 2532424 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
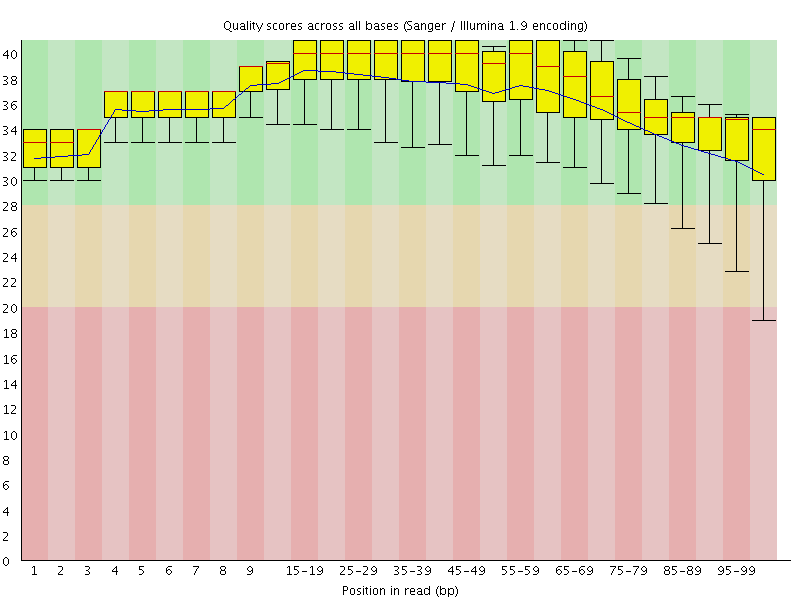
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
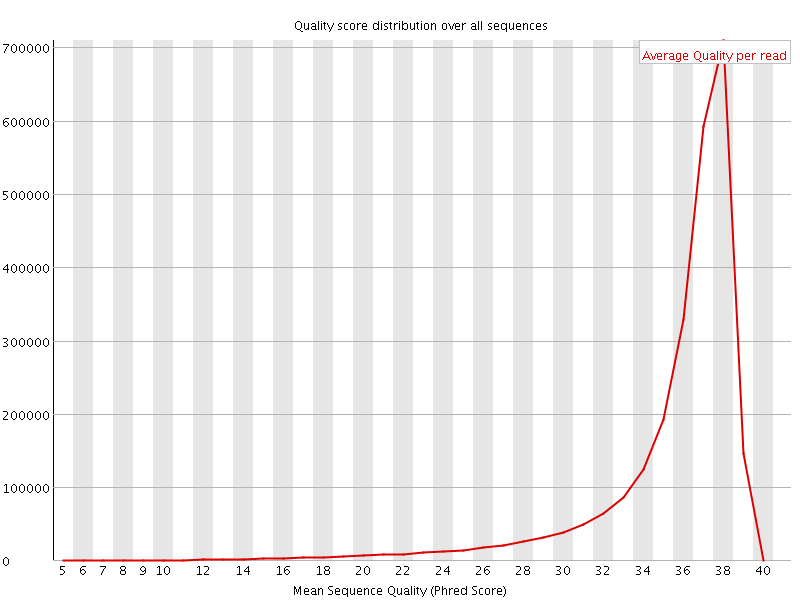
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
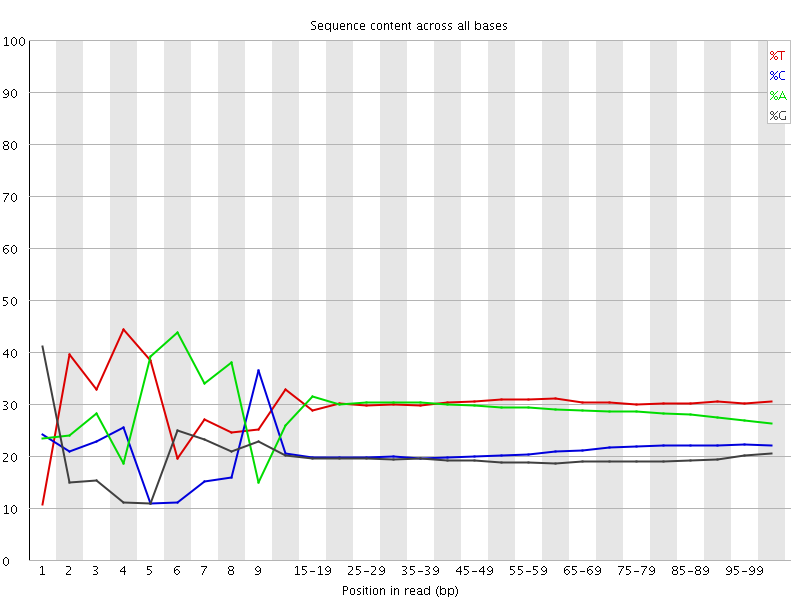
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
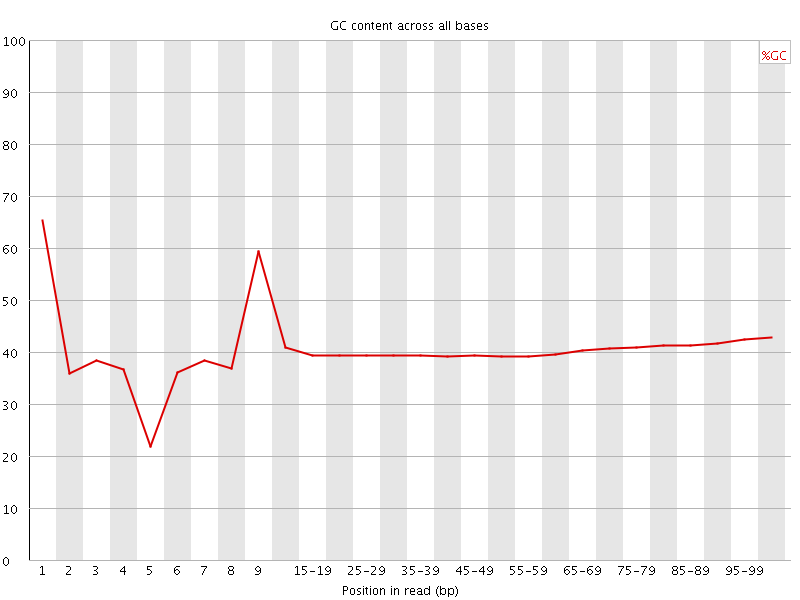
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
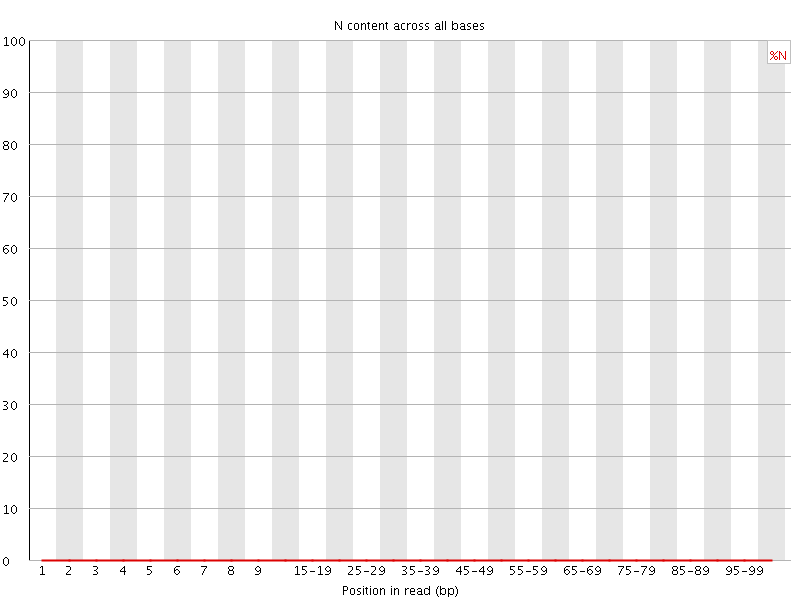
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
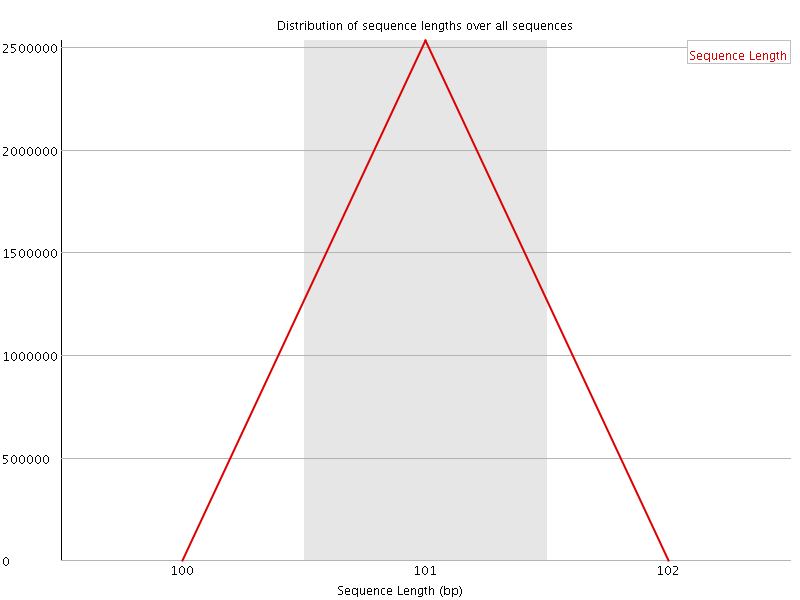
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
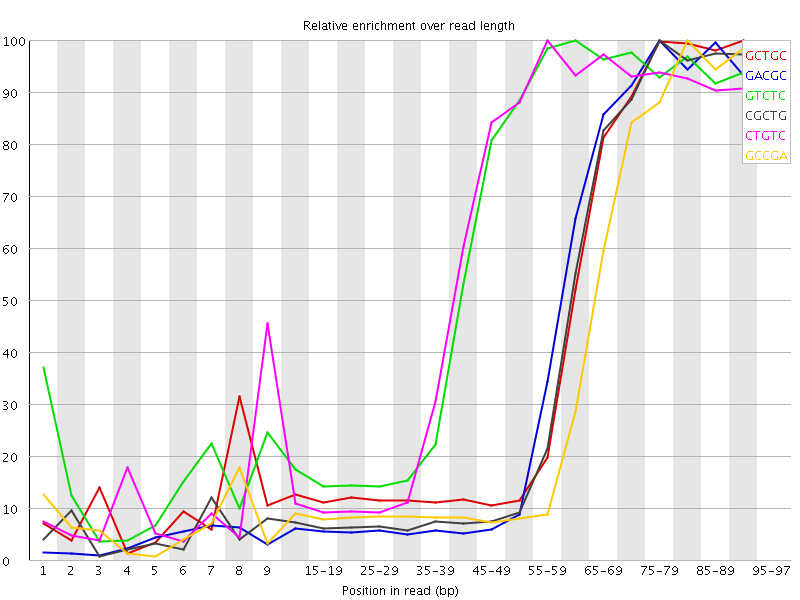
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCTGC | 548665 | 4.4066644 | 10.454717 | 90-94 |
| GACGC | 513935 | 4.307813 | 10.650259 | 75-79 |
| GTCTC | 830525 | 4.289414 | 7.1062574 | 60-64 |
| CGCTG | 511865 | 4.111101 | 10.366297 | 75-79 |
| CTGTC | 789490 | 4.077481 | 6.948232 | 55-59 |
| GCCGA | 476135 | 3.990973 | 10.94504 | 80-84 |
| CGACG | 467050 | 3.9148226 | 10.95643 | 95-97 |
| CTGCC | 499290 | 3.7590435 | 9.71766 | 90-94 |
| TGCCG | 462405 | 3.713858 | 10.24437 | 80-84 |
| TCTCT | 1093350 | 3.6311738 | 5.54254 | 60-64 |
| CCGAC | 455270 | 3.5771694 | 10.03264 | 80-84 |
| CTCTT | 1016960 | 3.377471 | 5.2828884 | 60-64 |
| TGTCT | 935465 | 3.3143134 | 5.1798635 | 55-59 |
| ACACA | 866650 | 3.2716696 | 6.4769416 | 6 |
| TGACG | 557930 | 3.2081094 | 7.437915 | 85-89 |
| CTGAC | 581370 | 3.1336021 | 6.88864 | 95-97 |
| GACGA | 504130 | 3.0252268 | 8.080811 | 85-89 |
| TACAC | 802130 | 2.9015129 | 7.337584 | 5 |
| ATCTG | 782670 | 2.8939476 | 5.6721406 | 70-74 |
| CACAT | 791550 | 2.8632424 | 5.132355 | 95-97 |
| ACGCT | 517260 | 2.7880473 | 6.9788013 | 75-79 |
| CATCT | 801120 | 2.7767153 | 5.155639 | 70-74 |
| ACATC | 763050 | 2.7601504 | 5.149686 | 70-74 |
| TCTGA | 739005 | 2.7324946 | 5.2711377 | 70-74 |
| AGAAG | 596185 | 2.5613158 | 5.3868136 | 5 |
| CCTTG | 456225 | 2.3562663 | 6.7233734 | 90-94 |
| CTCCT | 484000 | 2.3432164 | 6.518458 | 90-94 |
| ACTCC | 449610 | 2.2716897 | 6.478904 | 90-94 |
| TATAC | 837150 | 2.0773215 | 5.4897156 | 5 |
| CAAAG | 508215 | 2.0466871 | 5.1999764 | 4 |
| GGTGG | 210205 | 1.921331 | 7.6029973 | 95-97 |
| TTGTG | 506995 | 1.9162313 | 5.3125644 | 95-97 |
| AGAGA | 442555 | 1.9012942 | 5.4169593 | 8 |
| GATAC | 483870 | 1.867182 | 5.1237254 | 85-89 |
| ACGAT | 478295 | 1.8456689 | 5.1428146 | 85-89 |
| TGTGT | 484765 | 1.832211 | 5.0546966 | 95-97 |
| GAGAG | 281160 | 1.7998949 | 6.267929 | 7 |
| CTTTG | 504835 | 1.7886093 | 7.5838833 | 9 |
| AAGAG | 379130 | 1.6288092 | 5.0804844 | 5 |
| GTGTA | 405405 | 1.5991132 | 8.889223 | 1 |
| CTCGG | 194565 | 1.5626706 | 7.3027825 | 95-97 |
| CGGTG | 155090 | 1.3288157 | 6.9330673 | 95-97 |
| AAGAC | 320900 | 1.2923305 | 5.825066 | 5 |
| GAGTC | 215550 | 1.2394171 | 6.3492985 | 9 |
| TATGA | 430625 | 1.1399292 | 5.1809754 | 4 |
| GTGTT | 300000 | 1.1338756 | 5.6777973 | 1 |
| GTCTT | 319370 | 1.1315145 | 5.8808556 | 1 |
| GATTG | 286565 | 1.1303511 | 5.059528 | 7 |
| GACTT | 297290 | 1.0992393 | 5.4384923 | 7 |
| TCATA | 430430 | 1.068078 | 5.049161 | 2 |
| TGGAC | 174515 | 1.0034649 | 6.152679 | 5 |
| GTCCA | 174405 | 0.94004834 | 6.5597363 | 1 |
| AGACT | 240560 | 0.92828506 | 5.211582 | 6 |
| GGACT | 161180 | 0.9267884 | 5.844497 | 6 |
| GAGTA | 217060 | 0.89354396 | 5.177655 | 1 |
| GTATA | 262690 | 0.69538 | 7.1841145 | 1 |