![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-182_AAGAGGCA-ACTGCATA_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 2797648 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
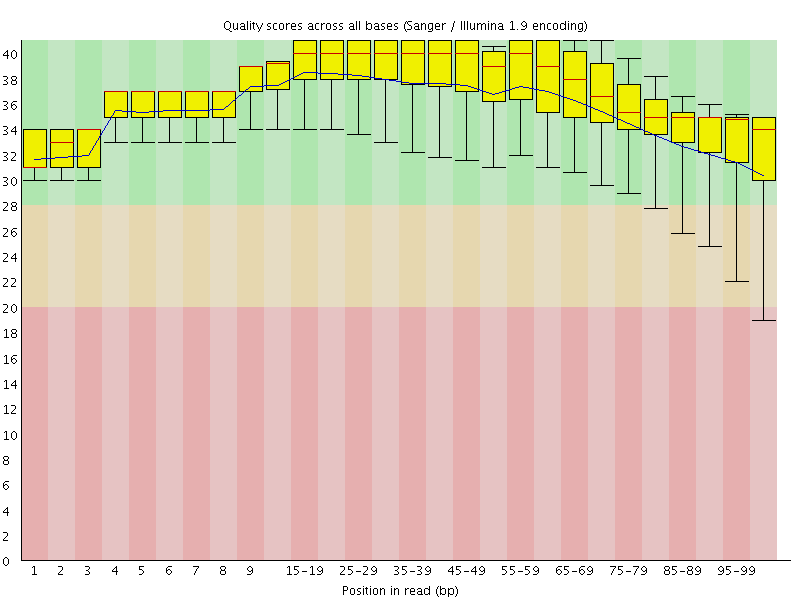
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
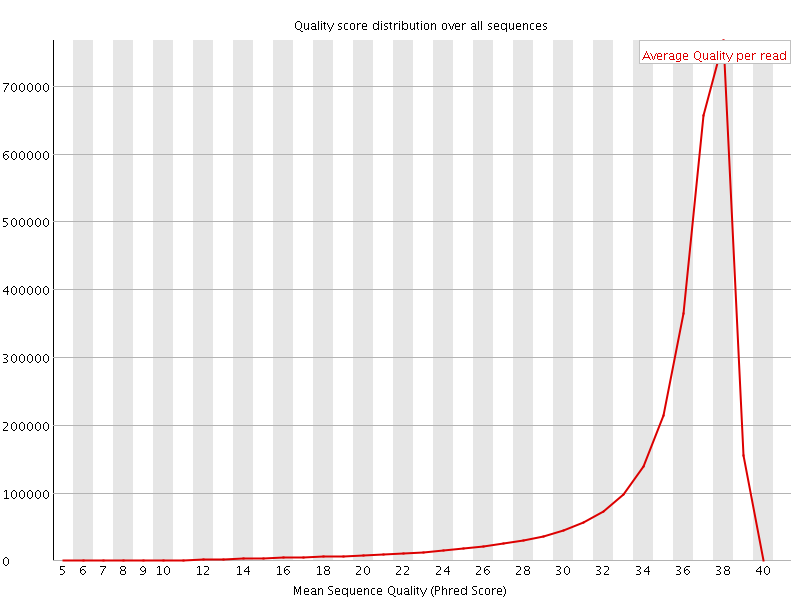
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
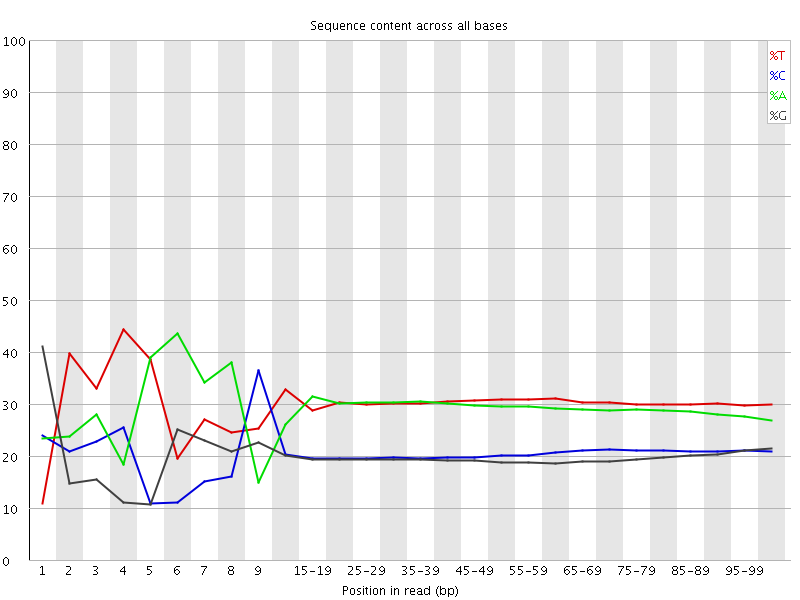
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
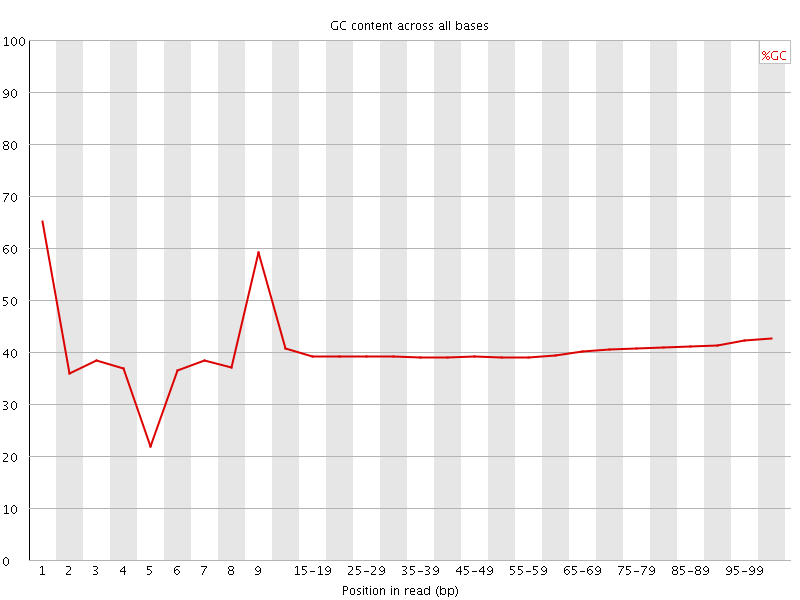
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
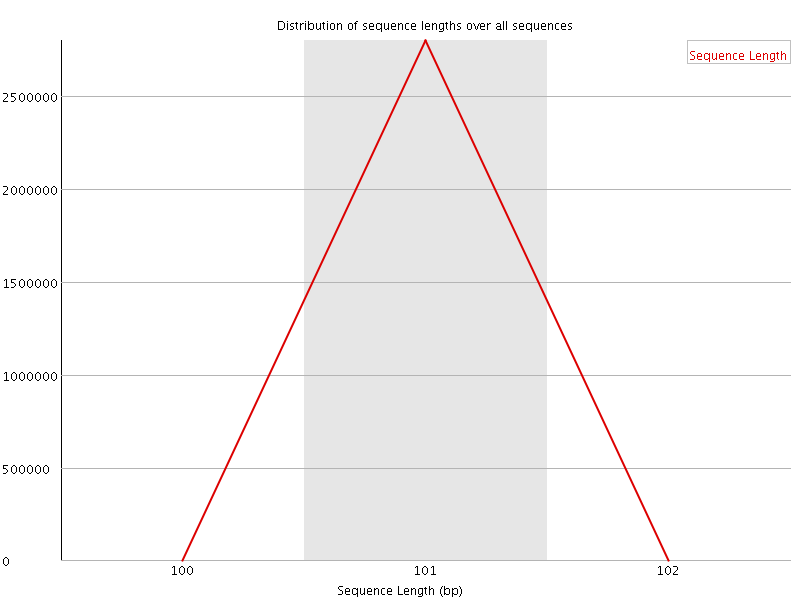
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
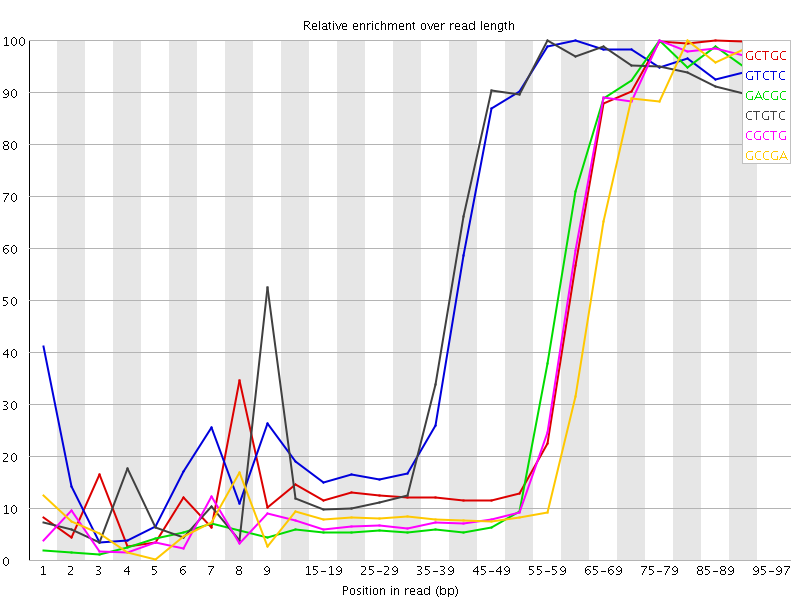
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCTGC | 580295 | 4.2947307 | 9.832515 | 85-89 |
| GTCTC | 874075 | 4.201662 | 6.775377 | 60-64 |
| GACGC | 533325 | 4.0879455 | 9.914435 | 75-79 |
| CTGTC | 824245 | 3.96213 | 6.5531697 | 55-59 |
| CGCTG | 532055 | 3.9377089 | 9.694186 | 75-79 |
| GCCGA | 493780 | 3.7848327 | 10.174417 | 80-84 |
| CTGCC | 522580 | 3.726267 | 9.444872 | 80-84 |
| CGACG | 485880 | 3.724279 | 10.082396 | 95-97 |
| TCTCT | 1188550 | 3.710866 | 5.495765 | 60-64 |
| TGCCG | 482255 | 3.5691416 | 9.654436 | 80-84 |
| CCGAC | 475770 | 3.5135357 | 9.6142235 | 80-84 |
| CTCTT | 1096400 | 3.4231572 | 5.1644506 | 60-64 |
| TGTCT | 984375 | 3.1899524 | 4.897417 | 55-59 |
| ACACA | 911390 | 3.1611154 | 5.9458675 | 6 |
| CTGAC | 610955 | 3.0416396 | 6.505112 | 70-74 |
| TGACG | 585235 | 3.0240896 | 6.8295894 | 75-79 |
| GACGA | 536025 | 2.8686361 | 7.4131207 | 85-89 |
| ATCTG | 832970 | 2.795626 | 5.281649 | 70-74 |
| TACAC | 827645 | 2.7717505 | 7.151285 | 5 |
| ACGCT | 542190 | 2.699293 | 6.5747304 | 75-79 |
| AGAAG | 659270 | 2.463378 | 5.4122963 | 5 |
| TGCAG | 463800 | 2.3965974 | 7.1407194 | 90-94 |
| TATAC | 879220 | 1.9849921 | 5.262337 | 5 |
| GTGTG | 378040 | 1.9576803 | 6.649264 | 95-97 |
| AGAGA | 520675 | 1.9455144 | 5.2695227 | 8 |
| GCAGT | 375930 | 1.942546 | 6.594903 | 90-94 |
| CAGTG | 370450 | 1.9142292 | 6.5805902 | 95-97 |
| GGTGG | 236880 | 1.8886307 | 7.611639 | 95-97 |
| GAGAG | 330300 | 1.8346992 | 6.1255927 | 7 |
| CTTTG | 534625 | 1.7324985 | 7.0170193 | 9 |
| CTCGG | 211270 | 1.5635972 | 7.19 | 95-97 |
| GTGTA | 426015 | 1.4840221 | 8.246496 | 1 |
| CGGTG | 171485 | 1.3172828 | 6.862833 | 95-97 |
| AAGAC | 347055 | 1.2493963 | 5.530623 | 5 |
| GAGTC | 225315 | 1.164272 | 5.202347 | 9 |
| GTCTT | 348505 | 1.1293606 | 5.8023214 | 1 |
| GTGTT | 325035 | 1.0932502 | 5.4154034 | 1 |
| GTTCT | 335245 | 1.0863904 | 5.076049 | 1 |
| GACTT | 319645 | 1.0727972 | 5.0016537 | 7 |
| TGGAC | 188500 | 0.9740375 | 5.8253837 | 5 |
| GTCCA | 188800 | 0.9399408 | 6.2236776 | 1 |
| GGACT | 170370 | 0.8803542 | 5.2497435 | 6 |
| GTATA | 285435 | 0.6688585 | 6.745415 | 1 |