![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-181_AAGAGGCA-GTAAGGAG_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1653252 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
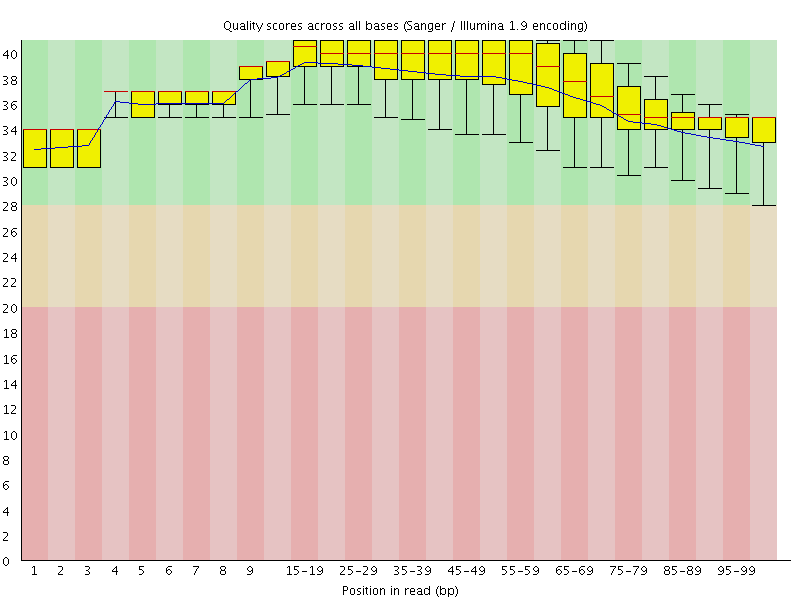
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
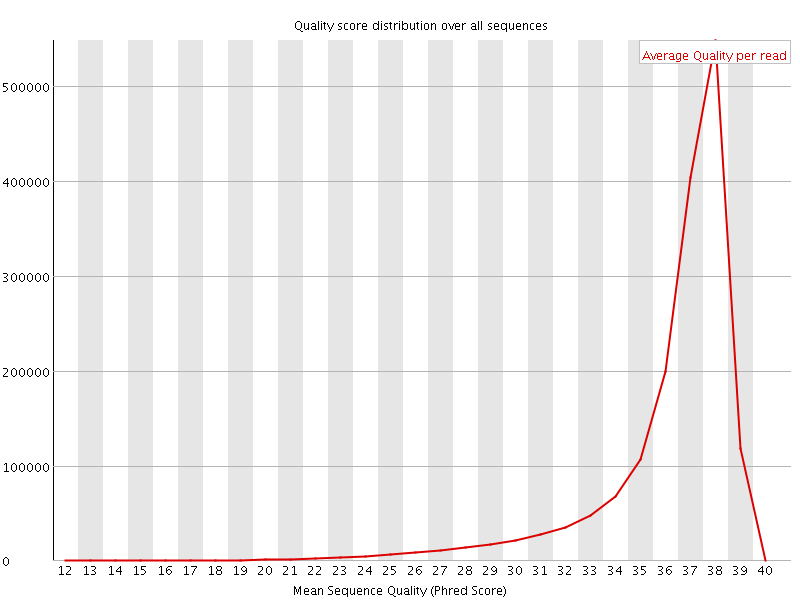
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
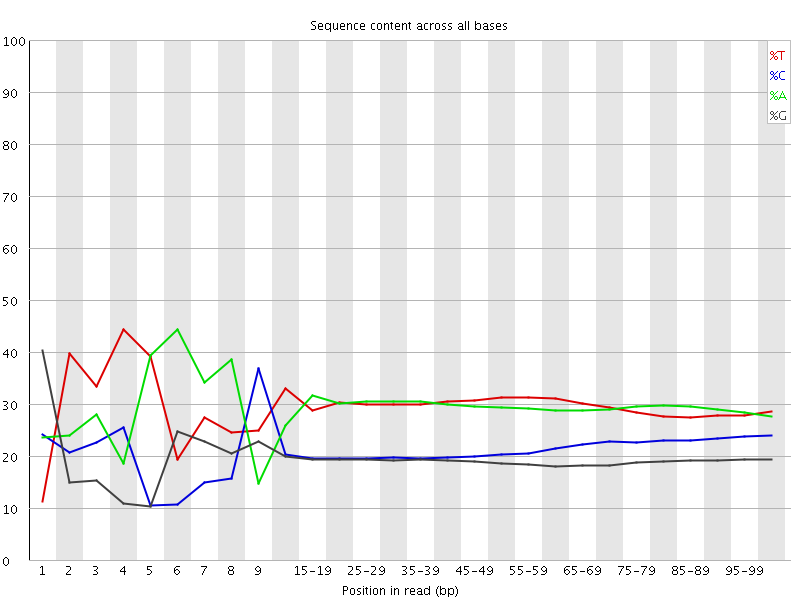
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
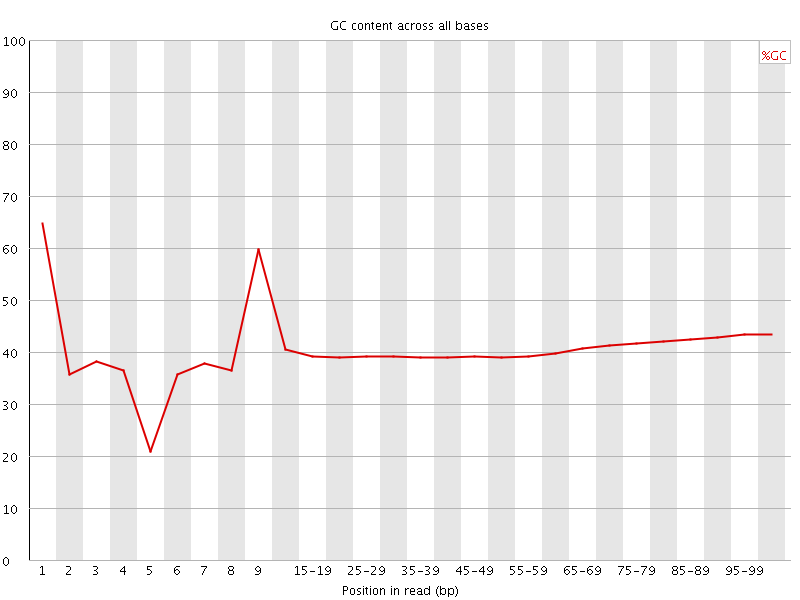
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
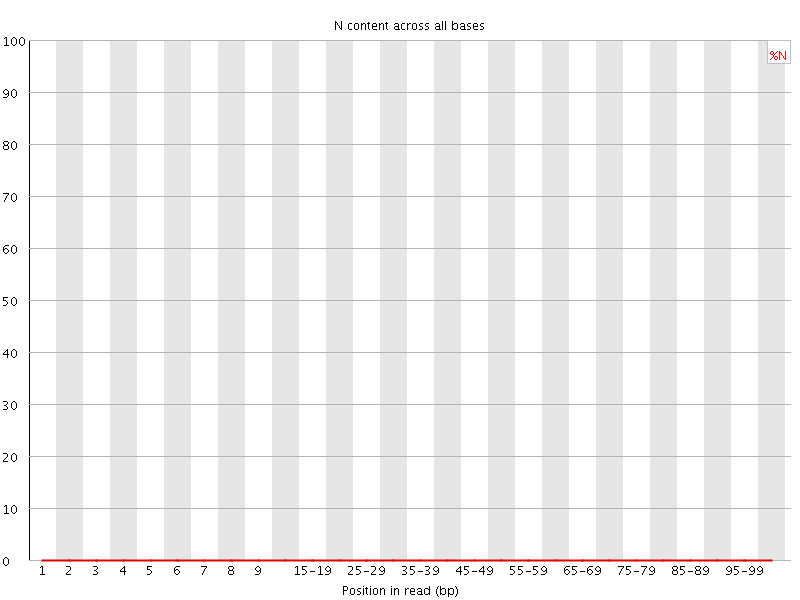
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
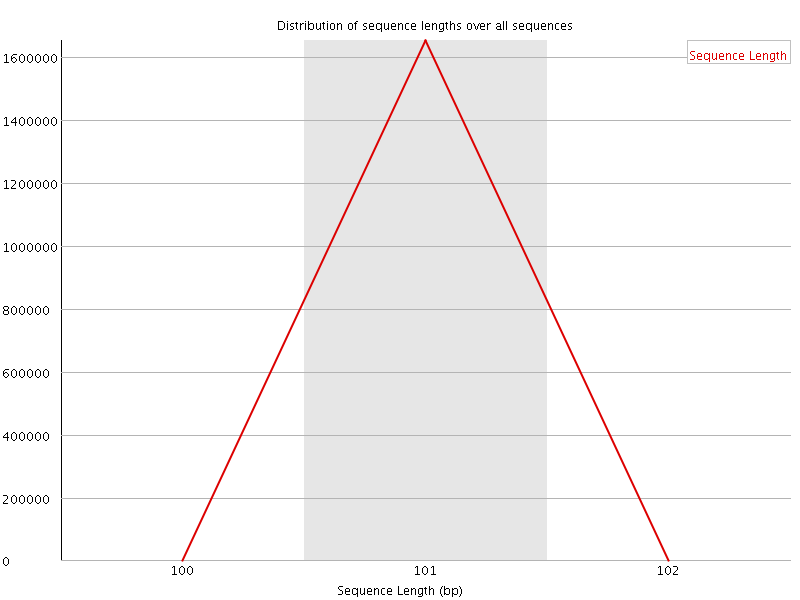
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
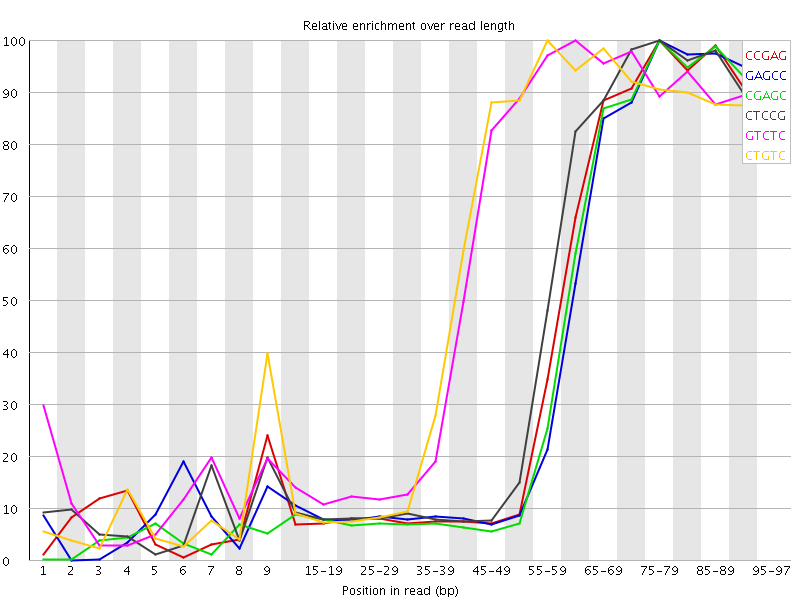
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCGAG | 431740 | 5.397214 | 13.034542 | 75-79 |
| GAGCC | 422170 | 5.2775784 | 13.124251 | 75-79 |
| CGAGC | 413205 | 5.165507 | 12.973929 | 75-79 |
| CTCCG | 454890 | 5.107055 | 11.575461 | 75-79 |
| GTCTC | 634870 | 5.089869 | 8.755805 | 60-64 |
| CTGTC | 612120 | 4.9074783 | 8.554531 | 55-59 |
| AGCCC | 402225 | 4.5509615 | 11.658127 | 80-84 |
| GCCCA | 396215 | 4.482962 | 11.863312 | 80-84 |
| TCTCT | 809430 | 4.1941643 | 6.7495646 | 60-64 |
| CCACG | 360985 | 4.0843534 | 11.627406 | 80-84 |
| TGTCT | 708415 | 4.055712 | 6.5531263 | 55-59 |
| TCTCC | 556755 | 4.039919 | 8.153735 | 70-74 |
| CCCAC | 383935 | 3.931684 | 10.641905 | 80-84 |
| CTCTT | 756880 | 3.9218705 | 6.412037 | 60-64 |
| ATCTC | 735235 | 3.8393912 | 10.253181 | 95-97 |
| TCCGA | 467835 | 3.7799363 | 8.61482 | 75-79 |
| GAGGC | 271710 | 3.7528934 | 14.363468 | 95-97 |
| GAGAC | 387265 | 3.484041 | 9.840299 | 85-89 |
| CGAGA | 385275 | 3.466138 | 9.719018 | 85-89 |
| ACACA | 647290 | 3.4330091 | 6.3737426 | 70-74 |
| AGAGG | 344390 | 3.423251 | 10.837244 | 90-94 |
| ACGAG | 375850 | 3.3813453 | 9.435618 | 95-97 |
| TACAC | 606135 | 3.1898887 | 7.351265 | 5 |
| CATCT | 603890 | 3.1535082 | 6.126196 | 70-74 |
| CACAT | 596040 | 3.136762 | 5.8227854 | 70-74 |
| CACGA | 372130 | 3.030096 | 8.534396 | 80-84 |
| AAGAG | 467640 | 3.02771 | 7.5994587 | 90-94 |
| ACATC | 574265 | 3.0221672 | 6.0024347 | 70-74 |
| CAAGA | 480770 | 2.8172584 | 6.9855227 | 90-94 |
| ACAAG | 450725 | 2.6411982 | 6.60441 | 85-89 |
| AGAAG | 397985 | 2.5767324 | 5.50297 | 5 |
| AGACA | 436730 | 2.559189 | 6.5794044 | 85-89 |
| AGGCA | 271670 | 2.444087 | 9.357098 | 95-97 |
| GACAA | 411700 | 2.4125159 | 6.4913154 | 85-89 |
| GGCAA | 266125 | 2.3942013 | 9.458884 | 95-97 |
| TATAC | 631860 | 2.3745682 | 5.761536 | 5 |
| CAAAG | 331255 | 1.9411173 | 5.091432 | 4 |
| AGAGA | 295880 | 1.915659 | 5.5312223 | 8 |
| TCTCG | 233610 | 1.872894 | 7.3770623 | 90-94 |
| CTTTG | 326590 | 1.8697445 | 8.147046 | 9 |
| GAGAG | 187260 | 1.8613721 | 6.4340158 | 7 |
| CAATC | 337810 | 1.7777826 | 5.8436007 | 95-97 |
| GCAAT | 290980 | 1.6919305 | 6.1275187 | 95-97 |
| CTCGT | 201415 | 1.6147809 | 7.234778 | 90-94 |
| GCTTT | 271035 | 1.5516894 | 5.291331 | 1 |
| GTATG | 234740 | 1.496407 | 6.1109433 | 95-97 |
| TTGAG | 227000 | 1.4470663 | 5.2389584 | 9 |
| GTCTT | 239355 | 1.3703197 | 6.0797553 | 1 |
| TGCCG | 105560 | 1.3094134 | 10.846568 | 95-97 |
| GCCGT | 104175 | 1.2922331 | 10.231059 | 95-97 |
| ATGCC | 156530 | 1.2647054 | 7.3757153 | 95-97 |
| GAGTC | 140255 | 1.2520549 | 6.4015603 | 9 |
| AAGAC | 213495 | 1.251057 | 5.7733216 | 5 |
| GATTG | 189200 | 1.2061013 | 5.9622135 | 7 |
| GTGTT | 187910 | 1.1886185 | 5.950547 | 1 |
| CCGTC | 105520 | 1.1846743 | 8.40706 | 95-97 |
| CCATG | 145205 | 1.1732033 | 5.041769 | 9 |
| TATGA | 280900 | 1.1663498 | 5.775908 | 4 |
| GTTCT | 197765 | 1.1322148 | 5.024821 | 1 |
| AGAAC | 192935 | 1.1305777 | 5.117004 | 5 |
| GACTT | 192725 | 1.1119555 | 5.7655406 | 7 |
| TATGC | 190795 | 1.1008202 | 5.382616 | 95-97 |
| TCGTA | 188605 | 1.0881846 | 5.24687 | 90-94 |
| TCATA | 282335 | 1.061032 | 5.16812 | 2 |
| TGGAC | 115245 | 1.0287908 | 6.938269 | 5 |
| GTGTA | 160750 | 1.0247397 | 9.888704 | 1 |
| CGTAT | 171715 | 0.9907353 | 5.268846 | 95-97 |
| GGACT | 104420 | 0.9321562 | 6.0985785 | 6 |
| GTCCA | 114655 | 0.9263706 | 6.441042 | 1 |
| AGACT | 159190 | 0.9256252 | 5.5398064 | 6 |
| CGTCT | 114280 | 0.9162037 | 5.4086876 | 95-97 |
| GAGTA | 141510 | 0.9091169 | 5.1464205 | 1 |
| GTATA | 168775 | 0.7007855 | 7.02692 | 1 |