![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-179_AAGAGGCA-TATCCTCT_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 2027320 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
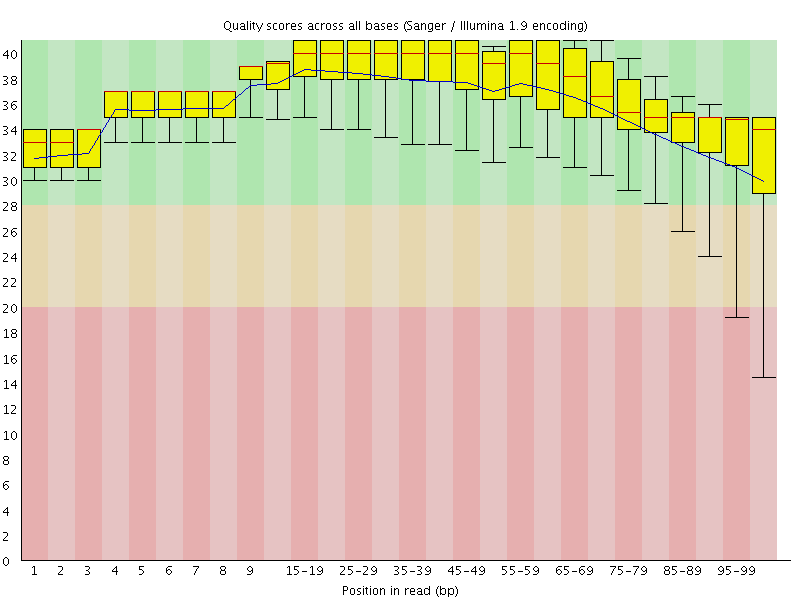
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
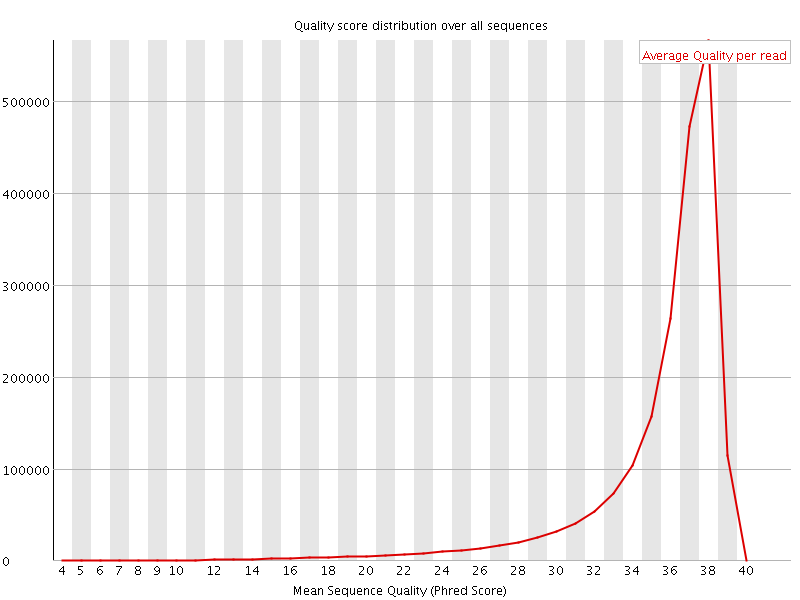
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
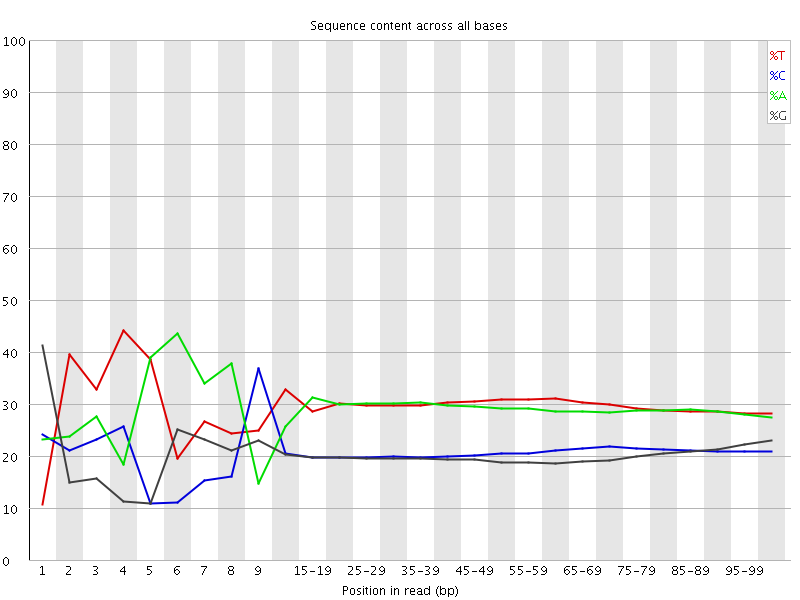
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content

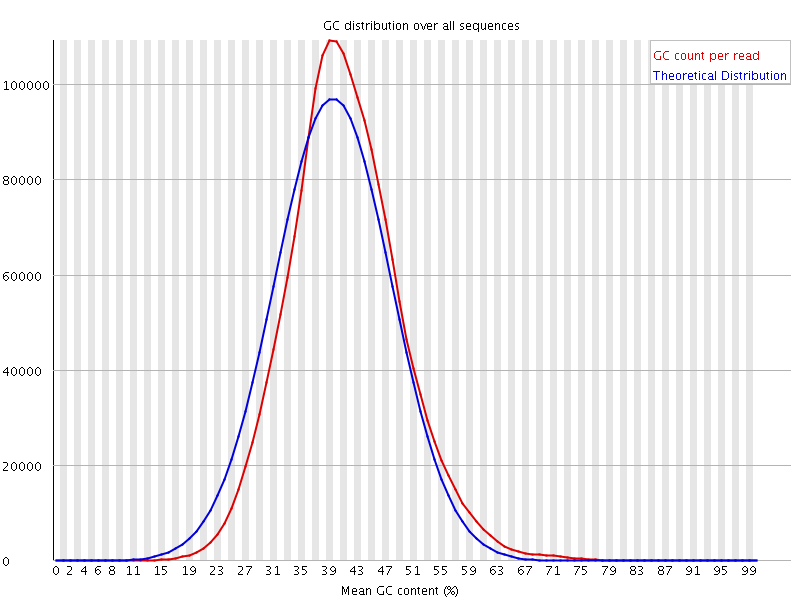
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
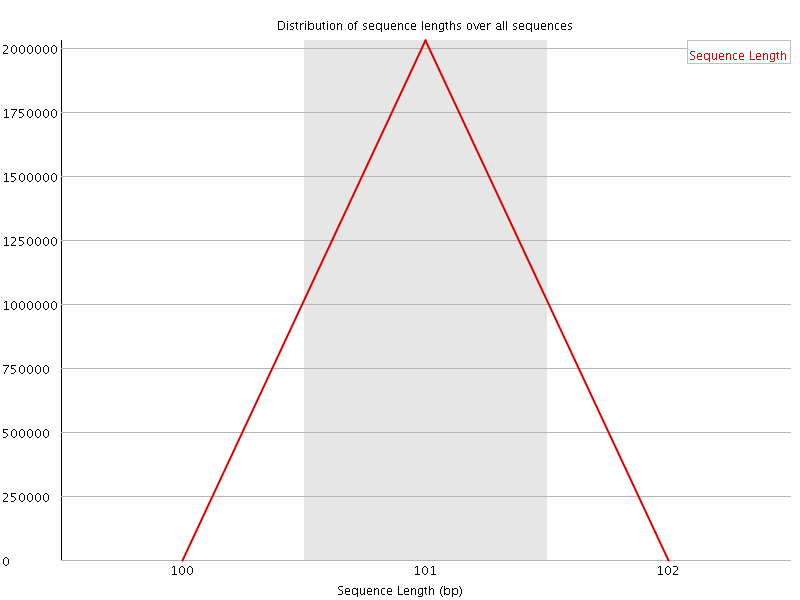
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
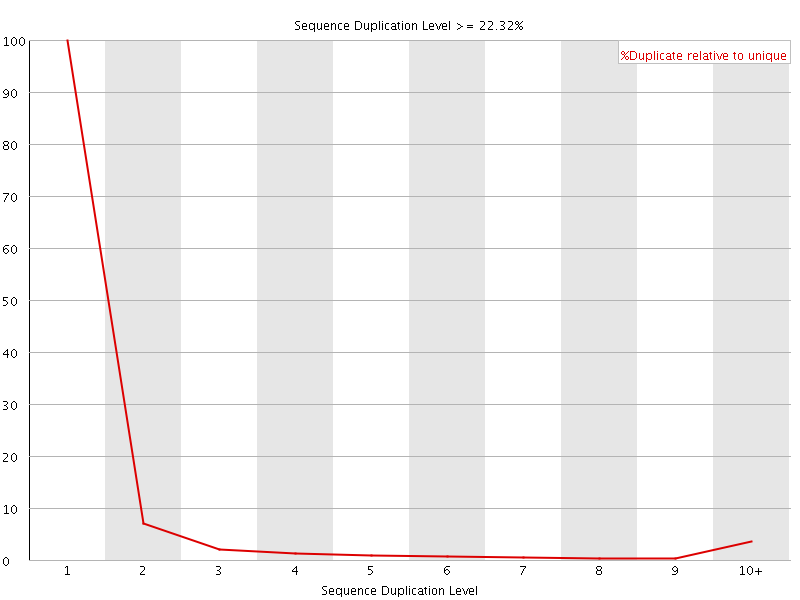
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
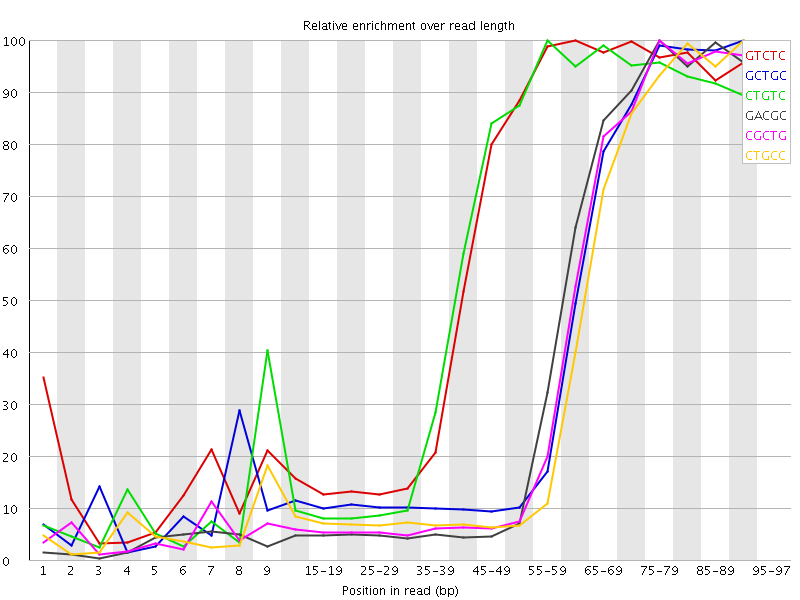
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GTCTC | 754745 | 4.951894 | 8.22797 | 60-64 |
| GCTGC | 497115 | 4.861105 | 11.85446 | 90-94 |
| CTGTC | 723360 | 4.745976 | 8.127379 | 55-59 |
| GACGC | 470225 | 4.7124696 | 11.840311 | 75-79 |
| CGCTG | 466550 | 4.562221 | 11.768413 | 75-79 |
| CTGCC | 452140 | 4.282814 | 11.428911 | 90-94 |
| TCTCT | 970070 | 4.2703857 | 6.667922 | 60-64 |
| GCCGA | 425615 | 4.2654 | 12.088325 | 80-84 |
| CGACG | 416595 | 4.1750035 | 12.20241 | 95-97 |
| TGCCG | 420280 | 4.1097636 | 11.586981 | 90-94 |
| CCGAC | 409340 | 3.9737923 | 11.526945 | 80-84 |
| CTCTT | 899990 | 3.9618835 | 6.308394 | 60-64 |
| TGTCT | 838645 | 3.8112202 | 6.091747 | 65-69 |
| GAAGA | 748980 | 3.7824404 | 7.9110336 | 85-89 |
| ACACA | 764445 | 3.6224673 | 6.6688285 | 6 |
| CTGAC | 527455 | 3.5466735 | 7.960355 | 85-89 |
| TGACG | 508310 | 3.5284693 | 8.384084 | 85-89 |
| GAGGA | 461500 | 3.3893464 | 9.386225 | 90-94 |
| TACAC | 712055 | 3.2923574 | 7.823556 | 5 |
| ATCTG | 703420 | 3.2761617 | 6.543204 | 70-74 |
| CACAT | 704050 | 3.255345 | 5.8916373 | 65-69 |
| CATCT | 713410 | 3.2186067 | 6.1128573 | 70-74 |
| GACGA | 447670 | 3.1847868 | 8.927835 | 95-97 |
| ACGCT | 473390 | 3.1831338 | 8.092367 | 75-79 |
| ACATC | 680990 | 3.1487215 | 6.0239754 | 70-74 |
| AGAGG | 426215 | 3.130206 | 9.257284 | 90-94 |
| TCTGA | 659340 | 3.07086 | 6.085997 | 70-74 |
| CGAAG | 414560 | 2.9492376 | 8.80505 | 85-89 |
| AAGAG | 570395 | 2.8805647 | 6.716363 | 90-94 |
| TATAC | 740345 | 2.3710592 | 5.378604 | 5 |
| ACGAA | 458305 | 2.241995 | 6.1381702 | 85-89 |
| AGGAT | 425360 | 2.096012 | 6.1801467 | 90-94 |
| CAAAG | 405950 | 1.9858782 | 5.2587686 | 4 |
| AGAGA | 365145 | 1.8440268 | 5.091754 | 8 |
| CTTTG | 405320 | 1.8419758 | 7.9824014 | 9 |
| GGATA | 372330 | 1.8347007 | 5.920132 | 90-94 |
| GGTGG | 169195 | 1.7632315 | 6.845715 | 95-97 |
| GAGAG | 236990 | 1.7405008 | 5.68432 | 7 |
| GATAG | 339125 | 1.671079 | 5.681967 | 95-97 |
| TAGTG | 342460 | 1.6465781 | 5.3435783 | 95-97 |
| GCTTT | 338465 | 1.5381534 | 5.178541 | 1 |
| GTGTA | 315925 | 1.5189954 | 9.136179 | 1 |
| CTCGG | 151705 | 1.4834675 | 6.8519974 | 95-97 |
| AAGAC | 260760 | 1.2756191 | 5.7476254 | 5 |
| CGGTG | 119880 | 1.210171 | 5.873974 | 95-97 |
| GAGTC | 172205 | 1.1953729 | 5.9382095 | 9 |
| GTCTT | 258145 | 1.1731393 | 6.1463485 | 1 |
| GTGTT | 242365 | 1.137045 | 5.7875214 | 1 |
| AACAC | 238745 | 1.1313384 | 5.05496 | 5 |
| GACTT | 240145 | 1.1184696 | 5.628596 | 7 |
| GATTG | 230220 | 1.1069182 | 5.3605933 | 7 |
| TATGA | 334505 | 1.1059428 | 5.481967 | 4 |
| TCATA | 334435 | 1.0710753 | 5.4934907 | 2 |
| TGGAC | 139685 | 0.96963316 | 6.1522 | 5 |
| AGACT | 196990 | 0.9402849 | 5.2313967 | 6 |
| GTCCA | 138455 | 0.9309887 | 6.8174286 | 1 |
| GGACT | 127850 | 0.88747954 | 5.7462573 | 6 |
| GAGTA | 175140 | 0.86302334 | 5.089222 | 1 |
| GTATA | 200905 | 0.6642337 | 6.7603045 | 1 |