![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-178_AAGAGGCA-CTCTCTAT_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1430012 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 39 |
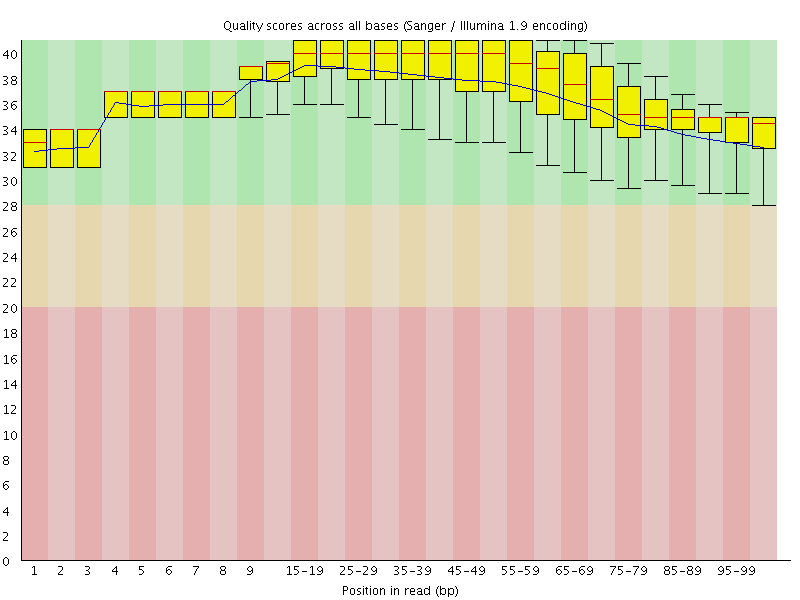
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
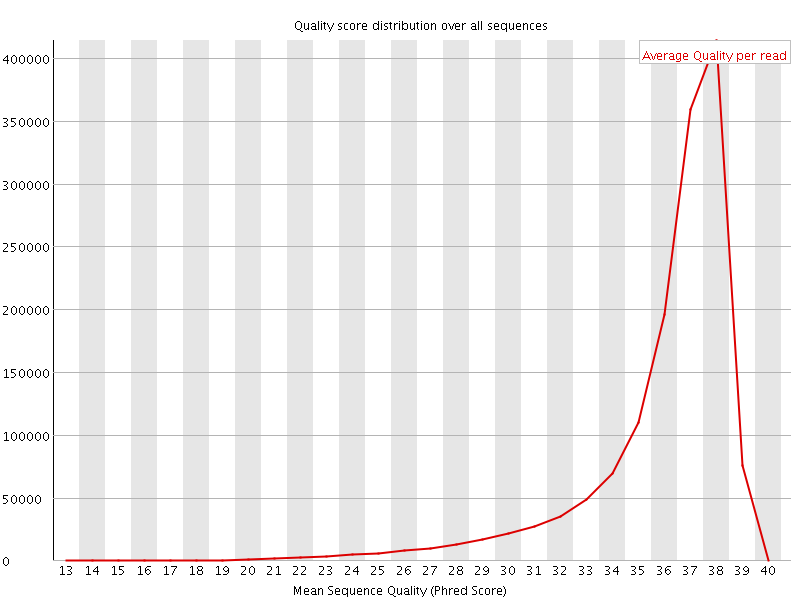
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
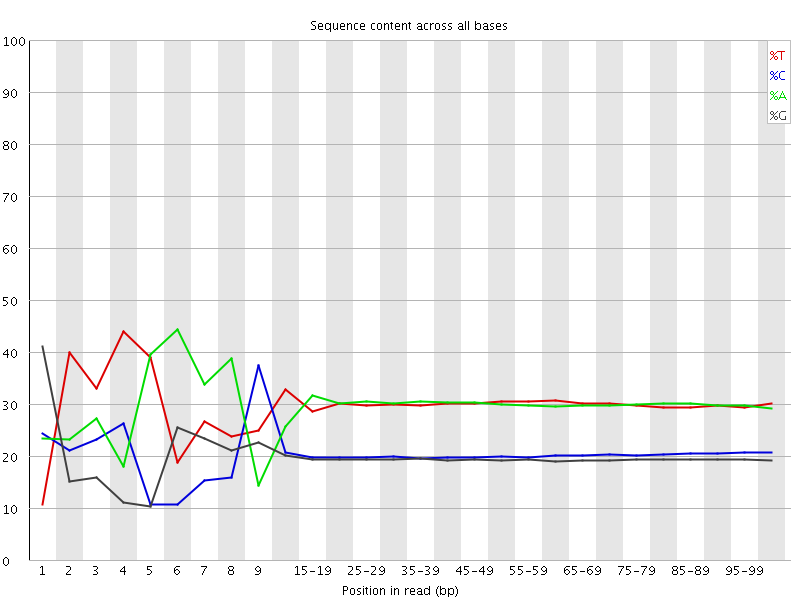
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
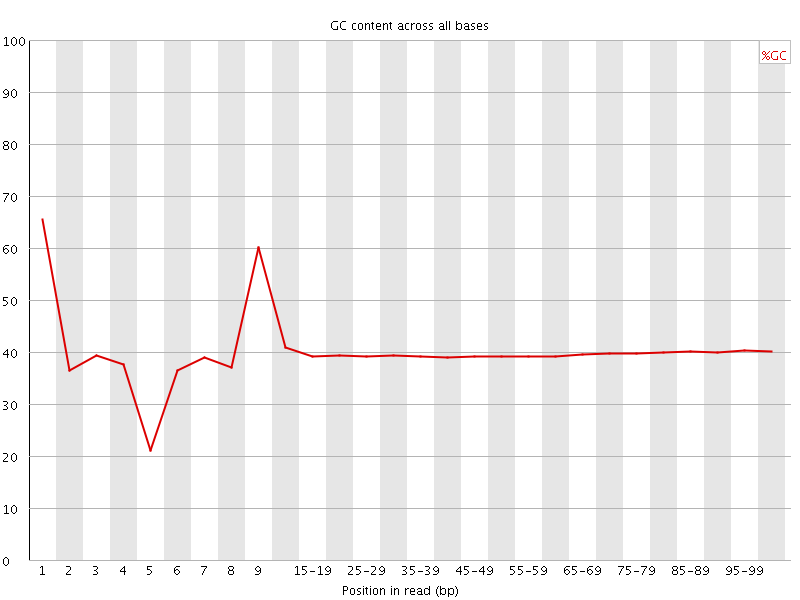
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
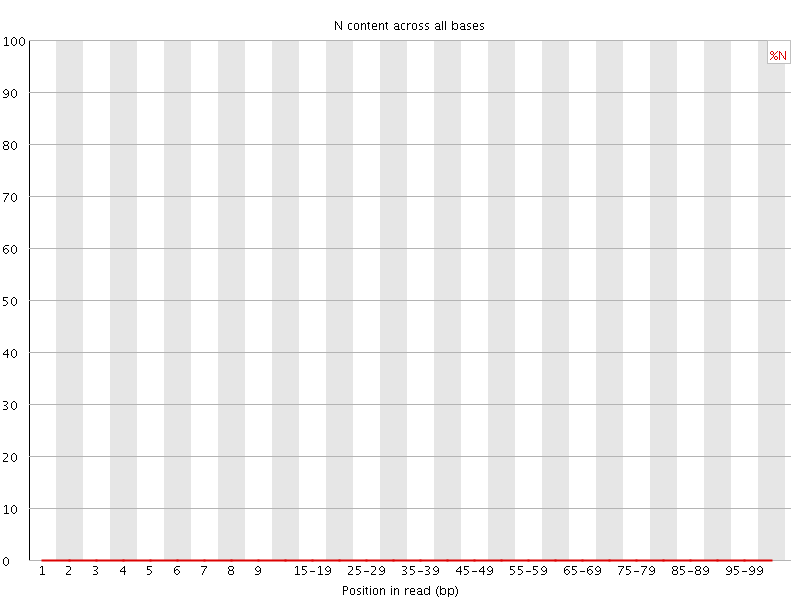
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
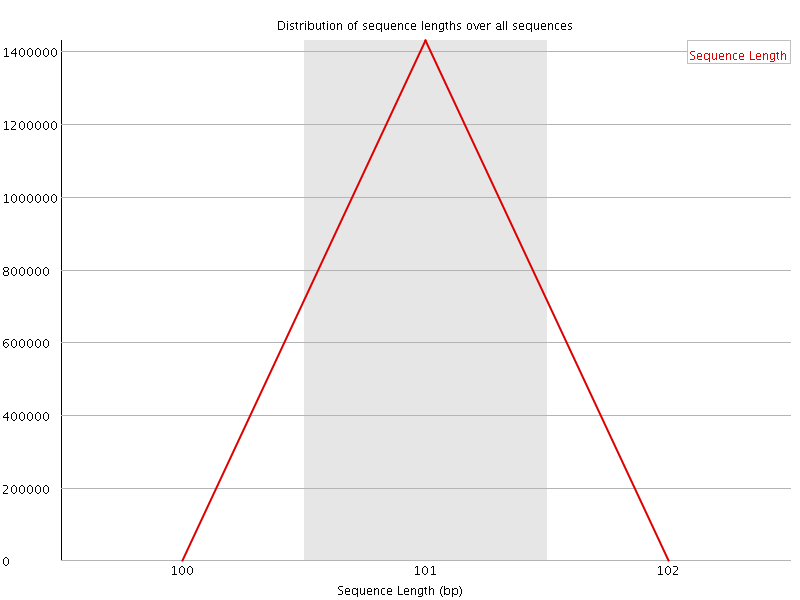
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
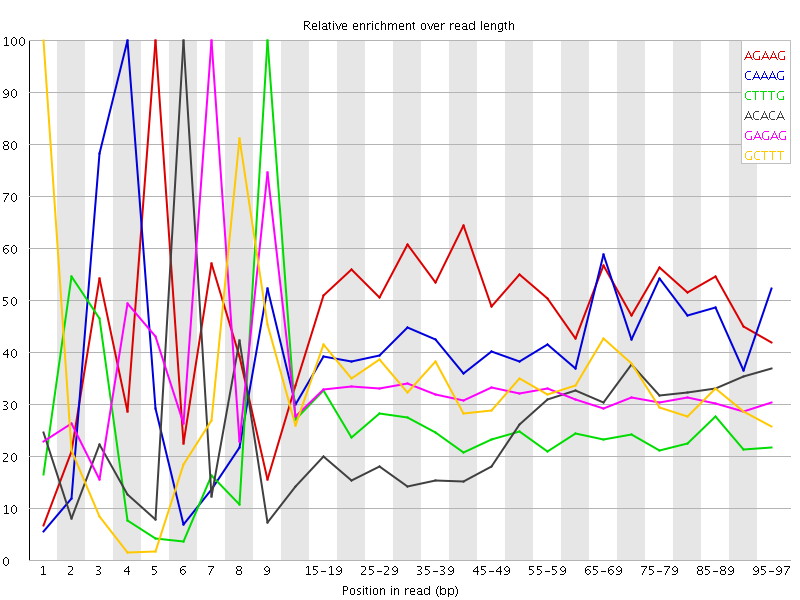
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AGAAG | 375490 | 2.6086402 | 5.197506 | 5 |
| CAAAG | 314255 | 2.113694 | 5.0482874 | 4 |
| CTTTG | 312420 | 2.0720682 | 8.280484 | 9 |
| ACACA | 298700 | 1.945088 | 7.662792 | 6 |
| GAGAG | 174380 | 1.8470111 | 5.6799064 | 7 |
| GCTTT | 261010 | 1.7311009 | 5.1907196 | 1 |
| CACAT | 255985 | 1.6591551 | 5.1381106 | 7 |
| TACAC | 254625 | 1.6503406 | 9.741984 | 5 |
| TTGAG | 218145 | 1.5014054 | 5.1257467 | 9 |
| ATACA | 325185 | 1.4279133 | 5.1118336 | 6 |
| GACTC | 140990 | 1.393214 | 5.442784 | 7 |
| CCATG | 140855 | 1.3918799 | 6.070429 | 9 |
| GAGTC | 134680 | 1.3746384 | 6.6214747 | 9 |
| GTCTT | 203770 | 1.3514671 | 6.2841797 | 1 |
| AAGAC | 199120 | 1.3392906 | 5.9581537 | 5 |
| GATTG | 181005 | 1.2457855 | 5.8765883 | 7 |
| GTGTT | 181355 | 1.242369 | 6.0125465 | 1 |
| GACTT | 185380 | 1.2352636 | 6.225751 | 7 |
| TATGA | 273525 | 1.2347888 | 5.782847 | 4 |
| GTTCT | 184965 | 1.2267464 | 5.55735 | 1 |
| AGAAC | 181185 | 1.2186589 | 5.27657 | 5 |
| TCATA | 275545 | 1.2042934 | 5.9341235 | 2 |
| TATAC | 264470 | 1.1558892 | 6.2492485 | 5 |
| ACATG | 168640 | 1.128987 | 5.3168626 | 8 |
| TGGAC | 109720 | 1.1198792 | 7.4281263 | 5 |
| GTATG | 159570 | 1.0982568 | 5.366016 | 3 |
| GTGTA | 157850 | 1.0864187 | 10.426113 | 1 |
| GTCCA | 109890 | 1.0858946 | 7.245028 | 1 |
| ACACC | 112795 | 1.084164 | 5.3314385 | 6 |
| GGACT | 98285 | 1.0031657 | 6.60168 | 6 |
| ACACT | 151915 | 0.98463005 | 5.12554 | 6 |
| GAGTA | 141905 | 0.981255 | 5.693517 | 1 |
| AGACT | 146315 | 0.97952884 | 5.839461 | 6 |
| CATAC | 137905 | 0.893825 | 5.0029798 | 3 |
| ATACT | 173260 | 0.75724787 | 5.143074 | 6 |
| GTATA | 167105 | 0.75437117 | 7.39459 | 1 |