![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-176_CGAGGCTG-CTAAGCCT_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1485139 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 41 |
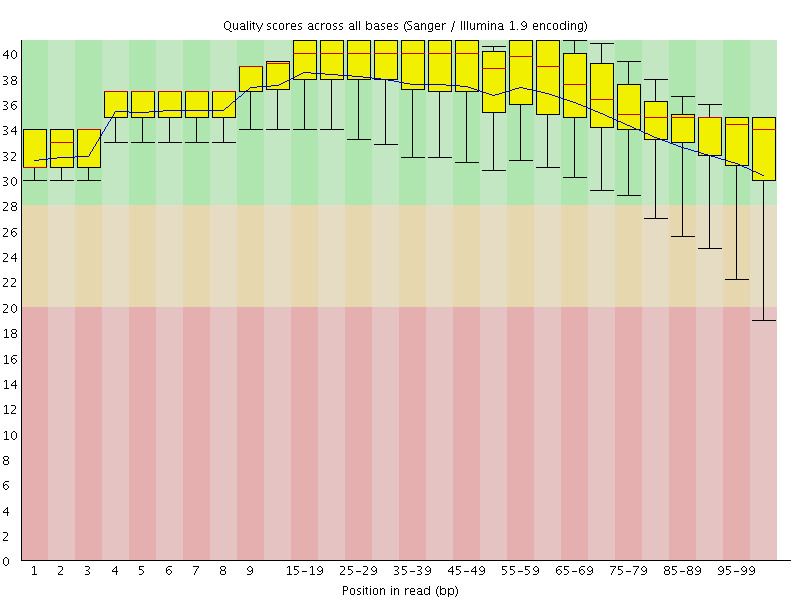
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
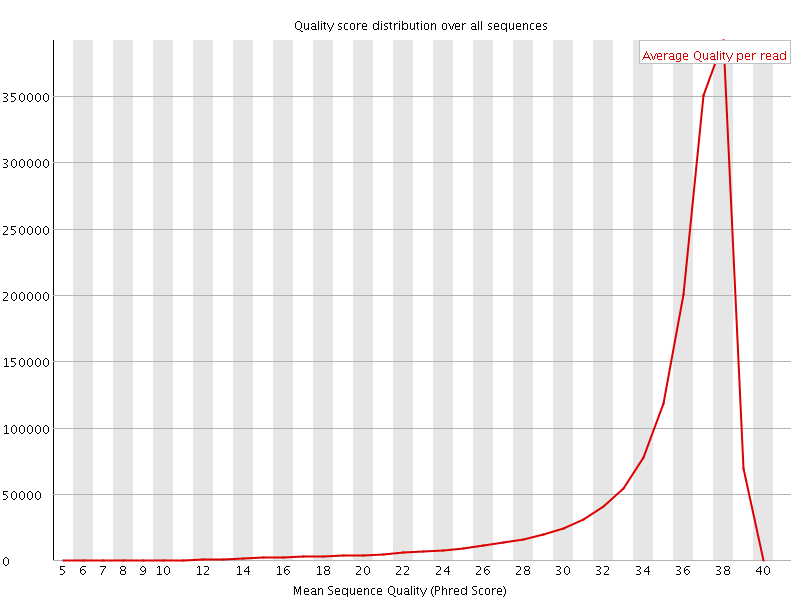
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
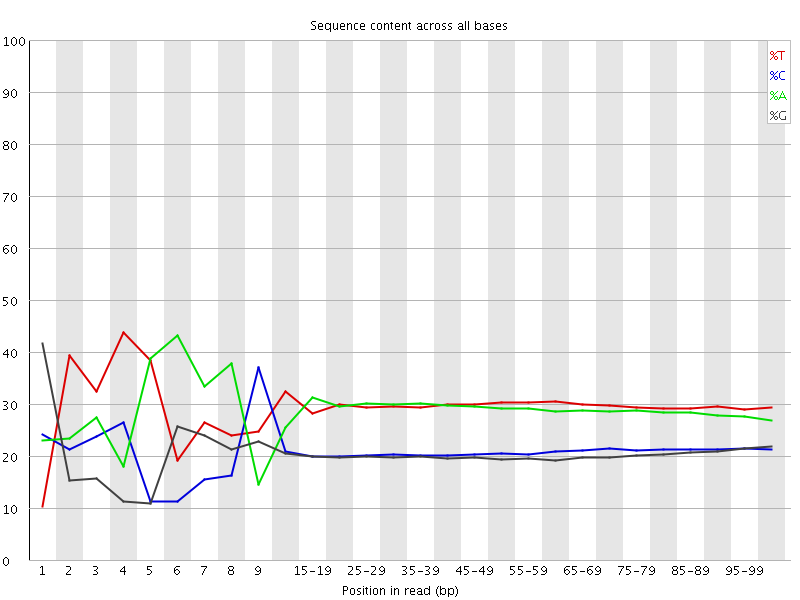
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
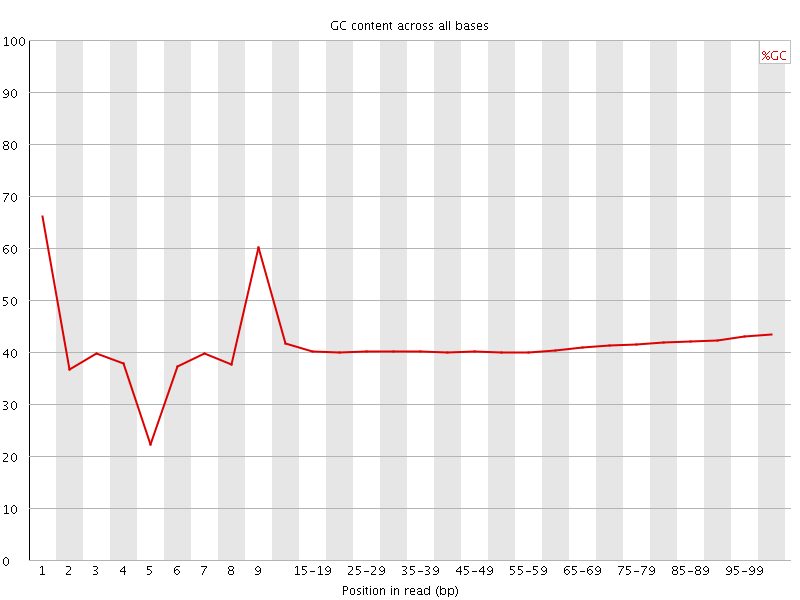
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
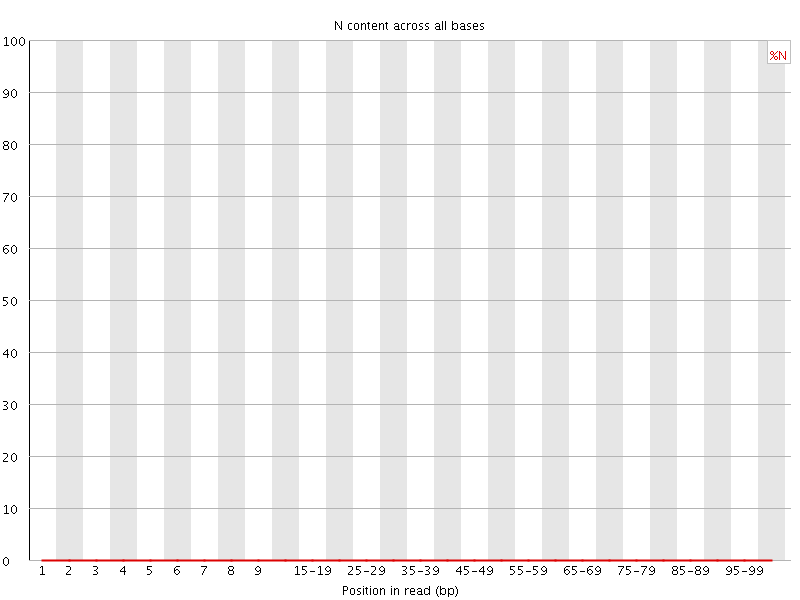
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
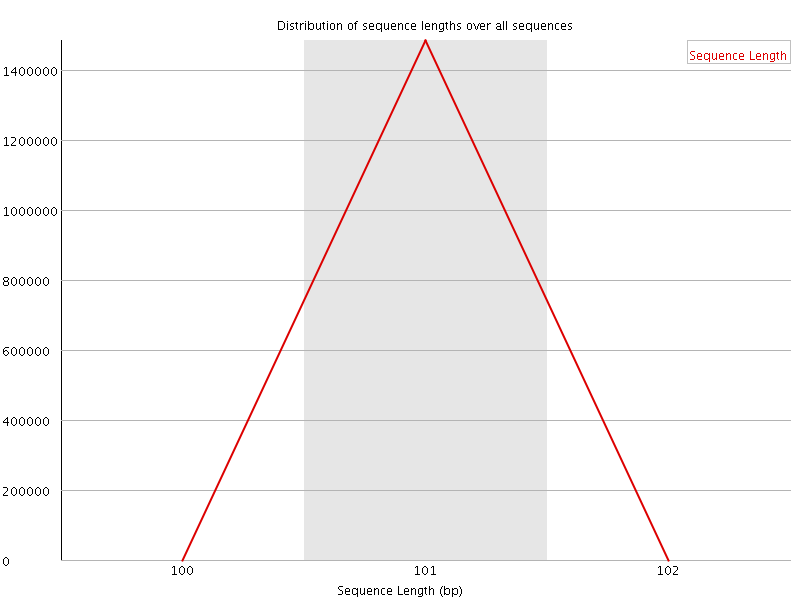
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
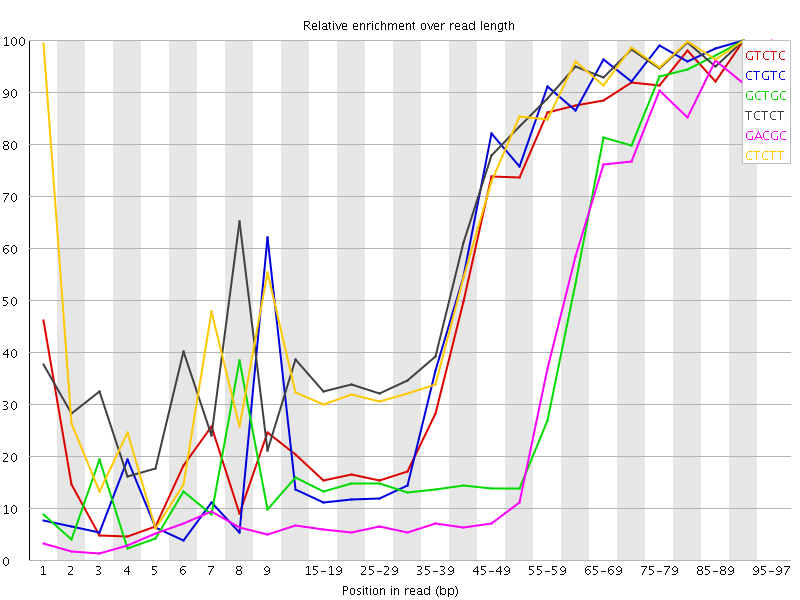
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GTCTC | 411200 | 3.6451871 | 6.228833 | 90-94 |
| CTGTC | 388245 | 3.4416971 | 5.775687 | 90-94 |
| GCTGC | 262360 | 3.415734 | 7.9268403 | 90-94 |
| TCTCT | 558640 | 3.371934 | 4.980638 | 90-94 |
| GACGC | 238285 | 3.1798773 | 8.295739 | 95-97 |
| CTCTT | 521305 | 3.146581 | 4.7359095 | 90-94 |
| CGCTG | 236115 | 3.0740435 | 7.7461863 | 85-89 |
| GCCGA | 223825 | 2.9869108 | 8.2001505 | 90-94 |
| CTGCC | 234625 | 2.9710922 | 7.7272162 | 90-94 |
| ACACA | 453115 | 2.9453514 | 7.165085 | 6 |
| CGACG | 220220 | 2.9388025 | 8.157657 | 95-97 |
| GAAGG | 286255 | 2.8181453 | 6.5545864 | 85-89 |
| CCGAC | 216090 | 2.804812 | 7.6699147 | 90-94 |
| TGCCG | 212520 | 2.766854 | 7.747517 | 90-94 |
| TACAC | 422160 | 2.677186 | 9.332877 | 5 |
| AGAAG | 379720 | 2.6090434 | 5.0992675 | 5 |
| CTGAC | 277020 | 2.5171268 | 6.0760255 | 95-97 |
| TGACG | 262515 | 2.4524074 | 6.1109605 | 95-97 |
| GGCTT | 260640 | 2.3754854 | 5.671583 | 95-97 |
| GACGA | 242845 | 2.3253853 | 6.293198 | 95-97 |
| ACGCT | 243690 | 2.2142756 | 5.519937 | 85-89 |
| CAAAG | 328835 | 2.1976132 | 5.040831 | 4 |
| CGAAG | 228075 | 2.1839538 | 6.070266 | 95-97 |
| ATACA | 479105 | 2.1801393 | 5.2652287 | 6 |
| AAGGC | 219660 | 2.1033752 | 5.4549065 | 90-94 |
| AGGCT | 222860 | 2.0819516 | 5.5048614 | 90-94 |
| CTTTG | 324750 | 2.0153048 | 8.30629 | 9 |
| TATAC | 427650 | 1.8985184 | 5.94554 | 5 |
| GGTGG | 128345 | 1.7662596 | 5.7203517 | 95-97 |
| GAGAG | 169415 | 1.6678699 | 5.4379244 | 7 |
| AGGTG | 165360 | 1.5882305 | 5.267251 | 95-97 |
| GTGTA | 230455 | 1.507126 | 10.382783 | 1 |
| TTGAG | 221650 | 1.4495431 | 5.2679234 | 9 |
| CTCGG | 107415 | 1.3984642 | 5.3717055 | 95-97 |
| AAGAC | 204785 | 1.3685839 | 5.893397 | 5 |
| GAGTC | 145090 | 1.3554265 | 7.167231 | 9 |
| CCATG | 146315 | 1.329483 | 5.411264 | 9 |
| TATGA | 276685 | 1.2628639 | 5.975474 | 4 |
| GTCTT | 203325 | 1.2617763 | 6.116735 | 1 |
| GACTT | 196840 | 1.2520802 | 6.4902825 | 7 |
| TCATA | 278530 | 1.236512 | 6.160838 | 2 |
| TGAGT | 187150 | 1.2239205 | 5.45827 | 8 |
| GTGTT | 184510 | 1.1772158 | 6.0411606 | 1 |
| GATTG | 178720 | 1.1687901 | 5.7467284 | 7 |
| GTTCT | 183545 | 1.1390272 | 5.297959 | 1 |
| ACACC | 123460 | 1.1184146 | 5.1794496 | 6 |
| ACATG | 165455 | 1.0787628 | 5.3279085 | 8 |
| TGGAC | 113380 | 1.0591927 | 6.774515 | 5 |
| GTCCA | 112970 | 1.0264955 | 7.3694415 | 1 |
| ACACT | 161585 | 1.0247136 | 5.301905 | 6 |
| AGACT | 152255 | 0.99269897 | 5.457328 | 6 |
| GGACT | 105580 | 0.9863253 | 6.2292314 | 6 |
| GAGTA | 146645 | 0.9830098 | 5.2870755 | 1 |
| GTATG | 140875 | 0.921292 | 5.0881557 | 3 |
| GTATA | 166405 | 0.7595166 | 7.2995186 | 1 |