![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-173_CGAGGCTG-GTAAGGAG_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 2438877 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 41 |
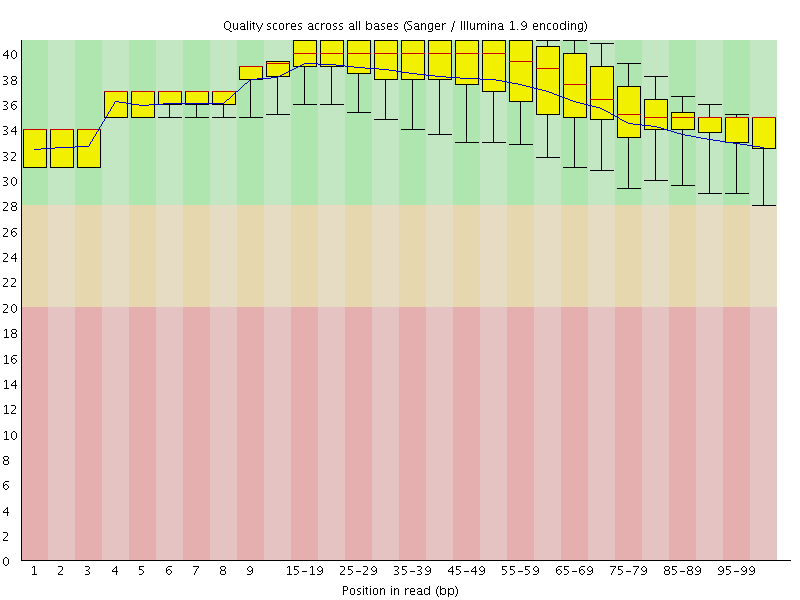
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
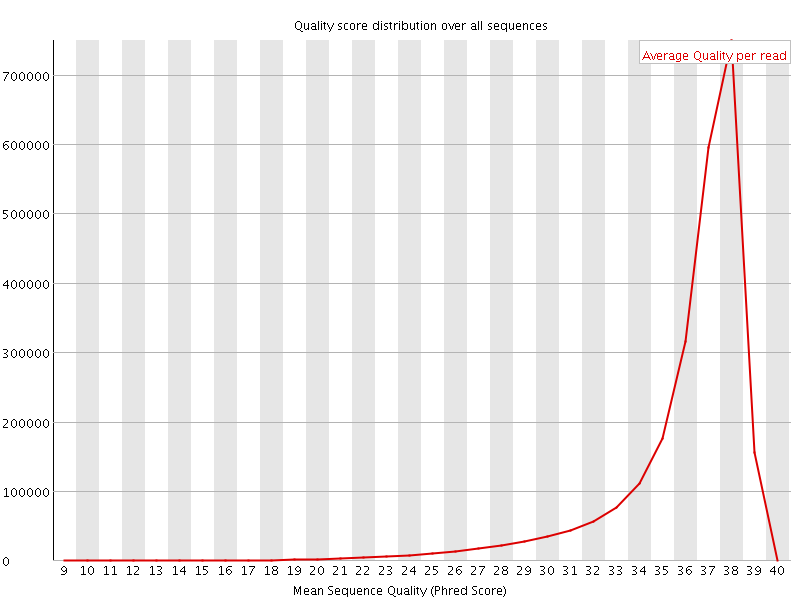
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
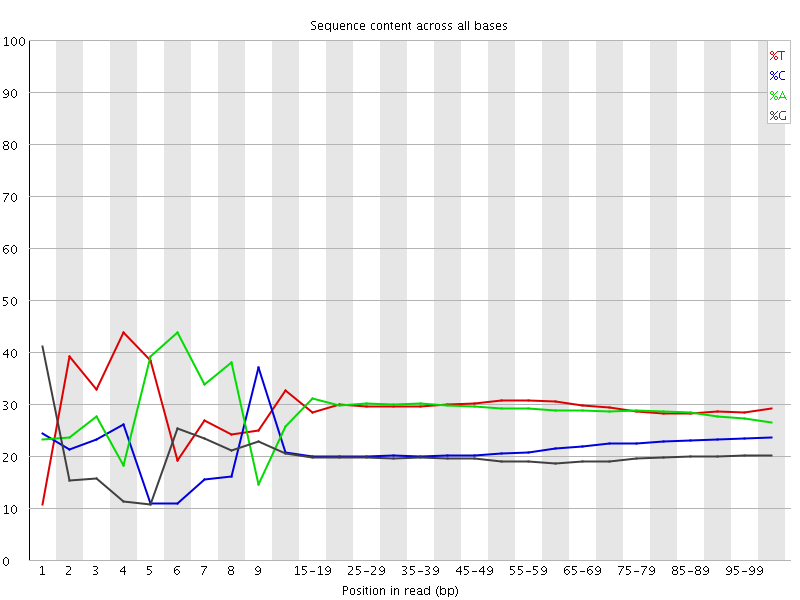
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
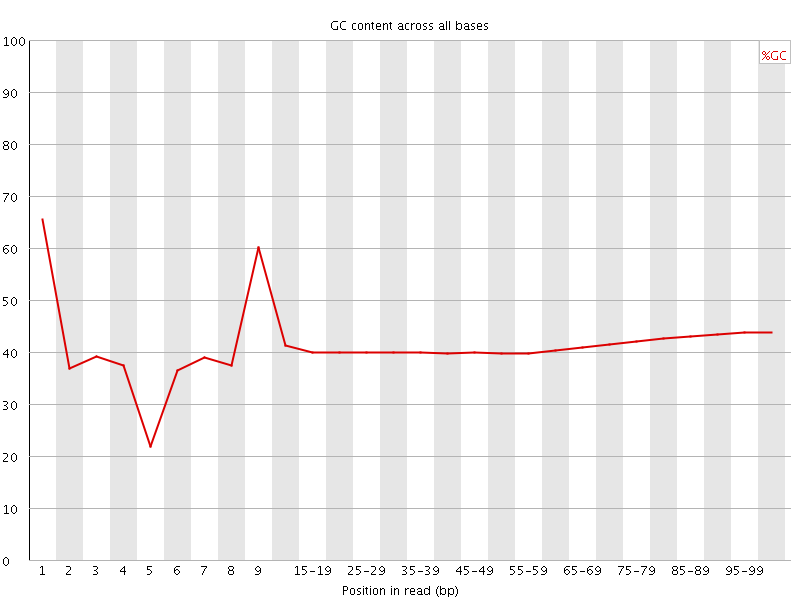
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
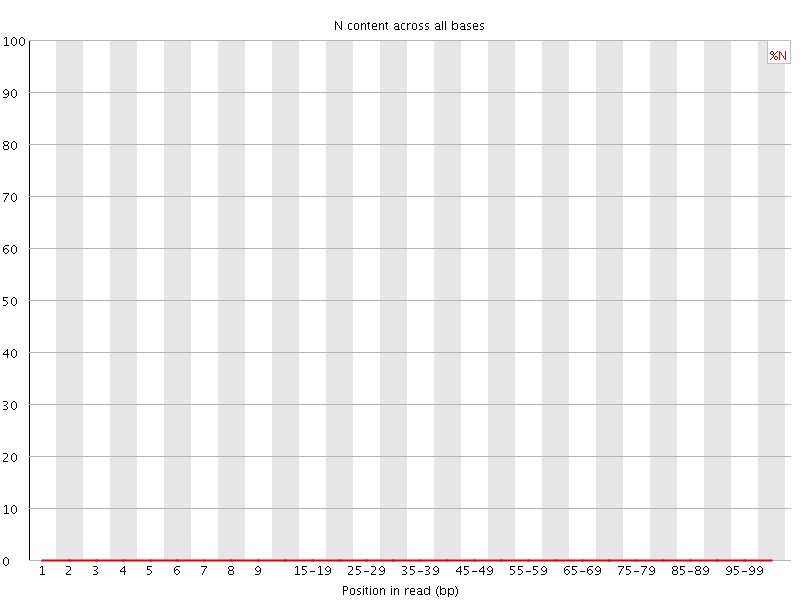
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
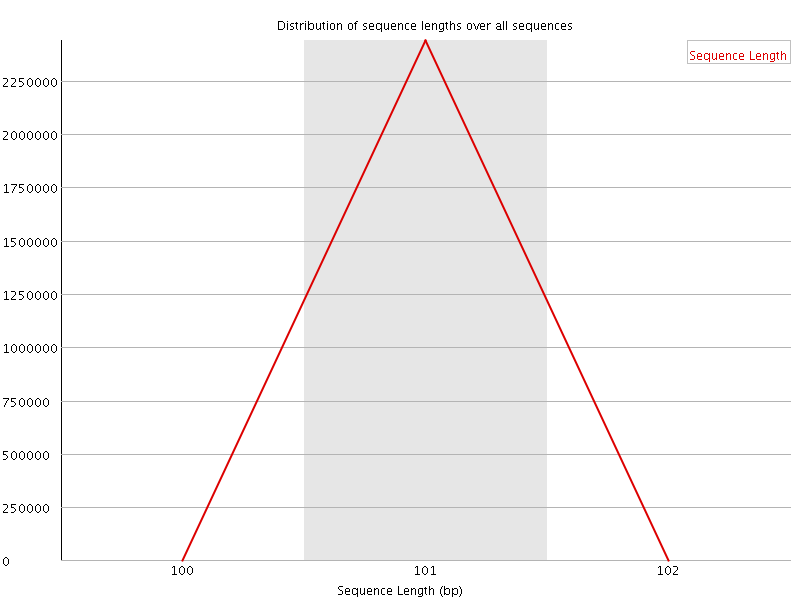
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
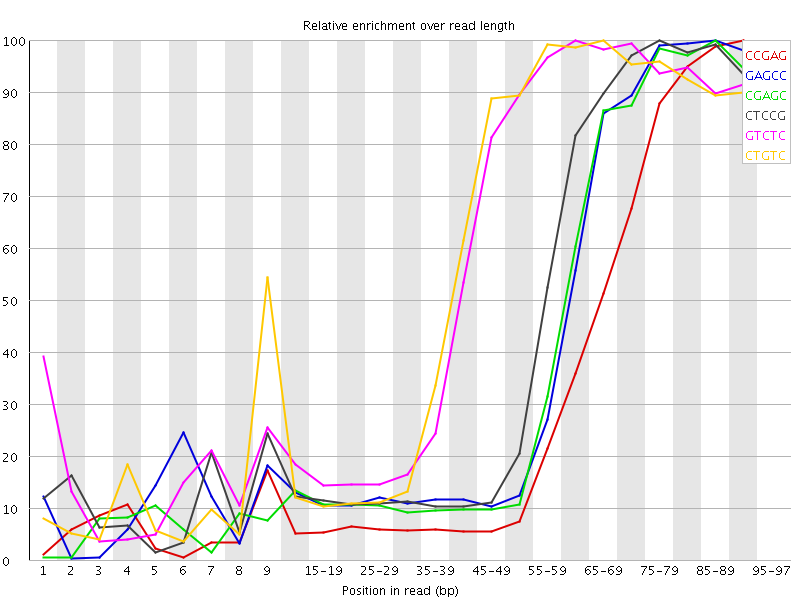
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCGAG | 775260 | 6.253706 | 17.774715 | 90-94 |
| GAGCC | 505725 | 4.079477 | 9.474255 | 85-89 |
| CGAGC | 498725 | 4.0230107 | 9.533708 | 85-89 |
| CTCCG | 541300 | 3.9639611 | 8.545114 | 75-79 |
| GTCTC | 746945 | 3.9461687 | 6.5274916 | 60-64 |
| CTGTC | 718395 | 3.7953365 | 6.3059044 | 65-69 |
| AGCCC | 484200 | 3.617589 | 8.688378 | 80-84 |
| GCCCA | 478490 | 3.574928 | 8.805025 | 80-84 |
| TCTCT | 990675 | 3.4971814 | 5.3780665 | 60-64 |
| TCTCC | 677935 | 3.3172596 | 6.3157096 | 80-84 |
| CTCTT | 927730 | 3.2749794 | 5.0782 | 60-64 |
| TGTCT | 842930 | 3.212729 | 5.0325727 | 65-69 |
| CCACG | 427895 | 3.196919 | 8.574894 | 90-94 |
| CCCAC | 461585 | 3.1941142 | 8.117207 | 90-94 |
| ATCTC | 879865 | 3.1688817 | 7.8629613 | 95-97 |
| GGCTG | 370635 | 3.1639464 | 10.438885 | 95-97 |
| CGAGG | 362070 | 3.153394 | 10.161399 | 90-94 |
| GAGGC | 351280 | 3.05942 | 10.27716 | 95-97 |
| TCCGA | 566695 | 3.0544946 | 6.570005 | 85-89 |
| ACACA | 791860 | 2.9685497 | 6.465338 | 6 |
| GACCG | 359520 | 2.9001012 | 9.161342 | 85-89 |
| CGAGA | 473765 | 2.8128827 | 7.3678 | 85-89 |
| GAGAC | 468855 | 2.7837307 | 7.3666487 | 85-89 |
| ACGAG | 453635 | 2.6933646 | 7.1663494 | 95-97 |
| TACAC | 730820 | 2.6853654 | 7.59486 | 5 |
| AGAAG | 576655 | 2.5200162 | 5.1954026 | 5 |
| CACGA | 443380 | 2.4381986 | 6.429491 | 95-97 |
| AGACC | 424035 | 2.3318183 | 6.603799 | 85-89 |
| AGGCT | 391740 | 2.2797308 | 7.1661963 | 95-97 |
| ACCGA | 378060 | 2.0789964 | 6.3812838 | 90-94 |
| GCTGA | 355345 | 2.0679302 | 6.9385786 | 95-97 |
| TATAC | 765045 | 2.0280354 | 5.5935683 | 5 |
| CAAAG | 488580 | 1.9775491 | 5.0926104 | 4 |
| CTTTG | 481215 | 1.8340946 | 7.9462557 | 9 |
| AGAGA | 416415 | 1.8197578 | 5.169976 | 8 |
| GAGAG | 265705 | 1.7032721 | 5.70959 | 7 |
| TCTCG | 302055 | 1.5957801 | 5.556261 | 90-94 |
| TTGAG | 335775 | 1.4097095 | 5.084933 | 9 |
| CTCGT | 263900 | 1.3942041 | 5.421005 | 90-94 |
| GTCTT | 338735 | 1.291049 | 5.8865466 | 1 |
| AAGAC | 303990 | 1.2304131 | 5.65584 | 5 |
| GAGTC | 208435 | 1.2129874 | 6.0664635 | 9 |
| TGCCG | 153225 | 1.2114812 | 7.710765 | 95-97 |
| TATGA | 418815 | 1.1986889 | 5.609085 | 4 |
| GCCGT | 151095 | 1.1946402 | 7.407905 | 95-97 |
| GATTG | 279480 | 1.1733619 | 5.571441 | 7 |
| ATGCC | 216710 | 1.1680703 | 5.5396705 | 95-97 |
| GTGTT | 279945 | 1.151996 | 5.5933223 | 1 |
| TCATA | 422410 | 1.1197543 | 5.512033 | 2 |
| GACTT | 287905 | 1.1195277 | 5.460038 | 7 |
| CCGTC | 151165 | 1.1069874 | 6.188924 | 95-97 |
| ACATG | 262235 | 1.0403495 | 5.043509 | 8 |
| TGGAC | 173265 | 1.0083157 | 6.941163 | 5 |
| GTGTA | 236520 | 0.9929997 | 9.271346 | 1 |
| GTCCA | 173770 | 0.9366229 | 6.403605 | 1 |
| GGACT | 155395 | 0.9043212 | 6.0213184 | 6 |
| GTATA | 248675 | 0.7117318 | 6.8297696 | 1 |