![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-167_GCTACGCT-AAGGAGTA_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 3243511 |
| Filtered Sequences | 0 |
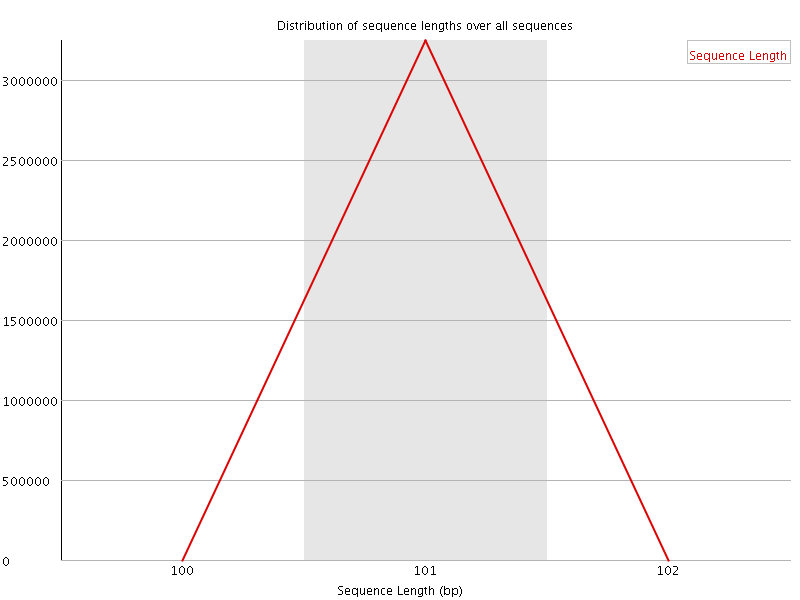
| Sequence length | 101 |
| %GC | 41 |
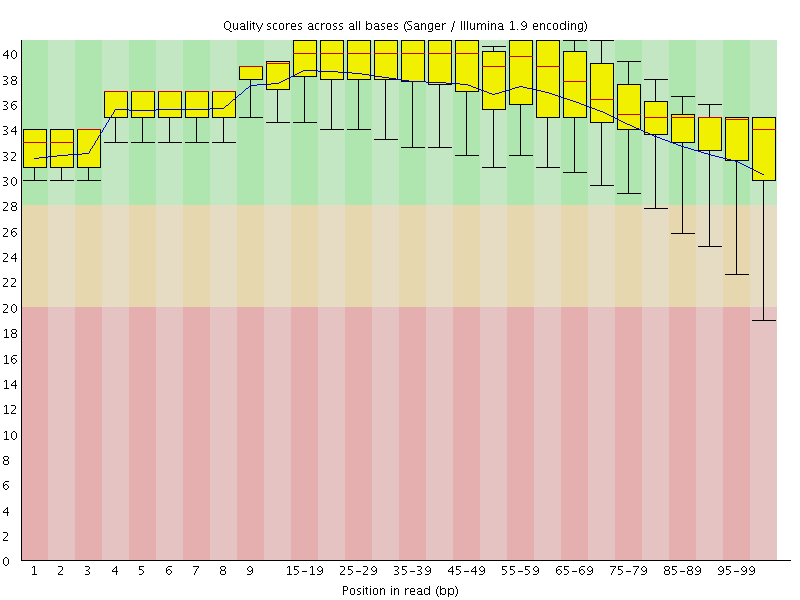
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
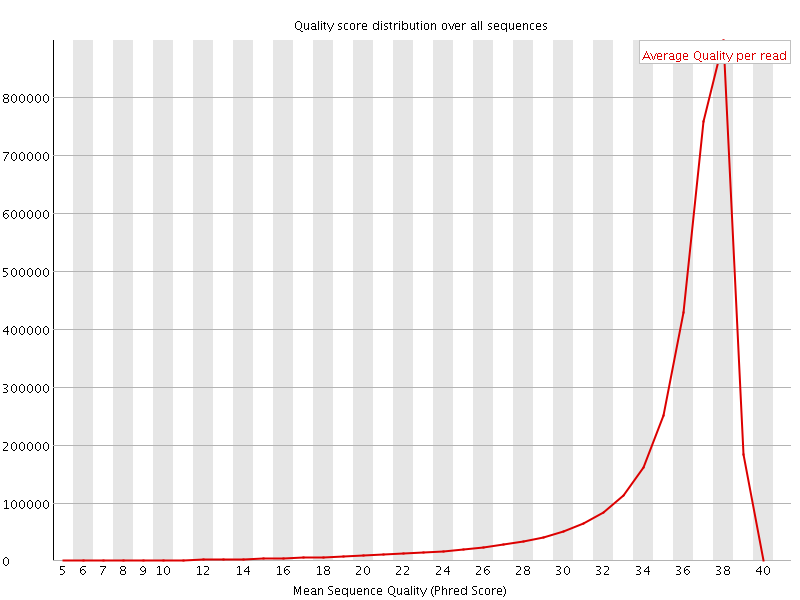
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
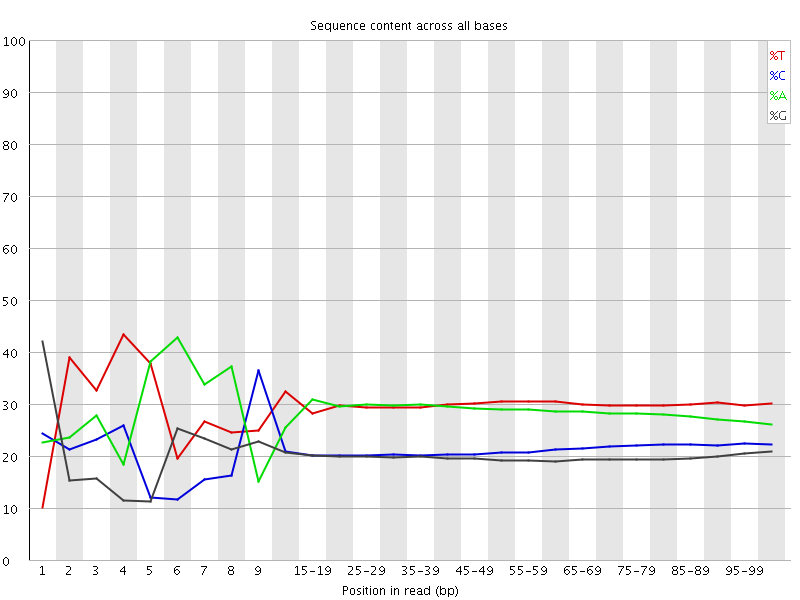
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
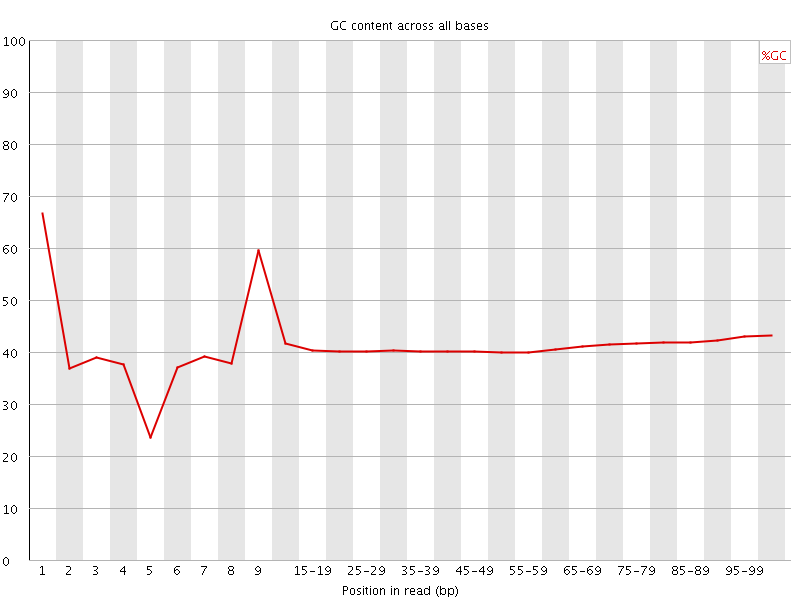
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
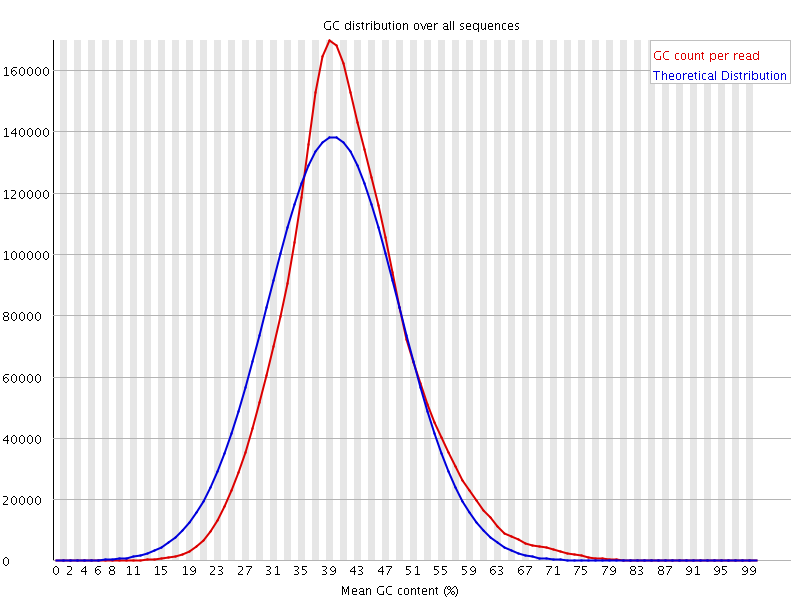
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels

![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
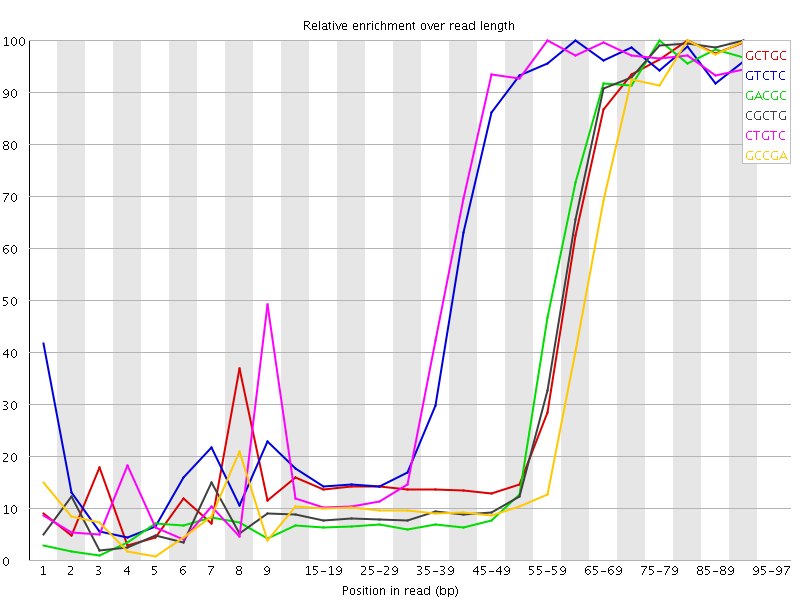
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCTGC | 720165 | 4.243607 | 9.44576 | 80-84 |
| GTCTC | 1024670 | 4.0212226 | 6.4708796 | 60-64 |
| GACGC | 652705 | 4.0107603 | 9.410377 | 75-79 |
| CGCTG | 660090 | 3.8896122 | 9.085527 | 90-94 |
| CTGTC | 982895 | 3.8572807 | 6.195886 | 55-59 |
| GCCGA | 613035 | 3.7669954 | 9.54315 | 80-84 |
| CGACG | 604180 | 3.712583 | 9.653611 | 95-97 |
| CTGCC | 649300 | 3.6067874 | 8.795087 | 80-84 |
| TGCCG | 595505 | 3.5090415 | 9.048446 | 80-84 |
| TCTCT | 1341130 | 3.5052252 | 5.2498755 | 60-64 |
| CCGAC | 581865 | 3.3705754 | 8.859735 | 95-97 |
| CTCTT | 1258805 | 3.2900577 | 4.9578214 | 60-64 |
| ACACA | 1088920 | 3.2274618 | 5.8714027 | 6 |
| TGTCT | 1158920 | 3.2131171 | 4.9241934 | 80-84 |
| TGACG | 702385 | 3.0491865 | 6.770767 | 75-79 |
| CTGAC | 733380 | 3.001303 | 6.365076 | 95-97 |
| GACGA | 646230 | 2.9255164 | 7.3989534 | 85-89 |
| TACAC | 1019160 | 2.8966813 | 6.8192677 | 5 |
| ATCTG | 966670 | 2.7948463 | 5.2157087 | 70-74 |
| ACGCT | 655720 | 2.6834853 | 6.2835402 | 75-79 |
| AGAAG | 784000 | 2.614792 | 5.076908 | 5 |
| ATACA | 1144795 | 2.397131 | 5.0101166 | 6 |
| CCTTG | 598695 | 2.3495233 | 6.1875052 | 95-97 |
| CTCCT | 620580 | 2.295852 | 5.913743 | 90-94 |
| ACTCC | 591750 | 2.2829218 | 5.941284 | 90-94 |
| CAAAG | 676170 | 2.12593 | 5.1245728 | 4 |
| TATAC | 1041640 | 2.0915825 | 5.1965733 | 5 |
| GGTGG | 295495 | 1.9593376 | 7.5310106 | 95-97 |
| CTTTG | 669390 | 1.8558902 | 7.987889 | 9 |
| GAGAG | 357465 | 1.7166309 | 5.5519695 | 7 |
| GTGTA | 542470 | 1.6637325 | 8.184284 | 1 |
| CTCGG | 271785 | 1.6015062 | 7.148975 | 95-97 |
| GCTTT | 570835 | 1.5826457 | 5.1376886 | 1 |
| CGCCG | 173660 | 1.4484518 | 5.283436 | 95-97 |
| CGGTG | 223445 | 1.3966957 | 6.880159 | 95-97 |
| AAGAC | 421265 | 1.3244894 | 5.6187277 | 5 |
| GAGTC | 295945 | 1.2847534 | 6.804238 | 9 |
| GTCTT | 417360 | 1.1571347 | 6.0334215 | 1 |
| GACTT | 400070 | 1.1566865 | 5.836917 | 7 |
| GTGTT | 389435 | 1.1453443 | 5.7139297 | 1 |
| TGAGT | 373315 | 1.1449413 | 5.644502 | 8 |
| GTTCT | 379635 | 1.0525417 | 5.0610266 | 1 |
| TGGAC | 233940 | 1.015578 | 6.345023 | 5 |
| GTCCA | 233420 | 0.95525396 | 6.8709006 | 1 |
| GGACT | 217720 | 0.94516385 | 6.172447 | 6 |
| AGACT | 311140 | 0.9380851 | 5.172838 | 6 |
| GTATA | 332540 | 0.7083196 | 6.847525 | 1 |