![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-165_GCTACGCT-GTAAGGAG_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 2604350 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
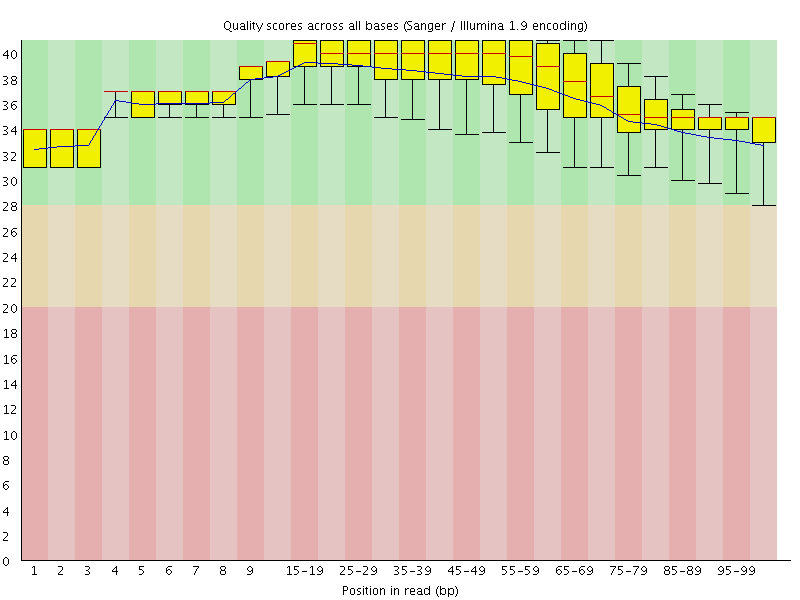
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
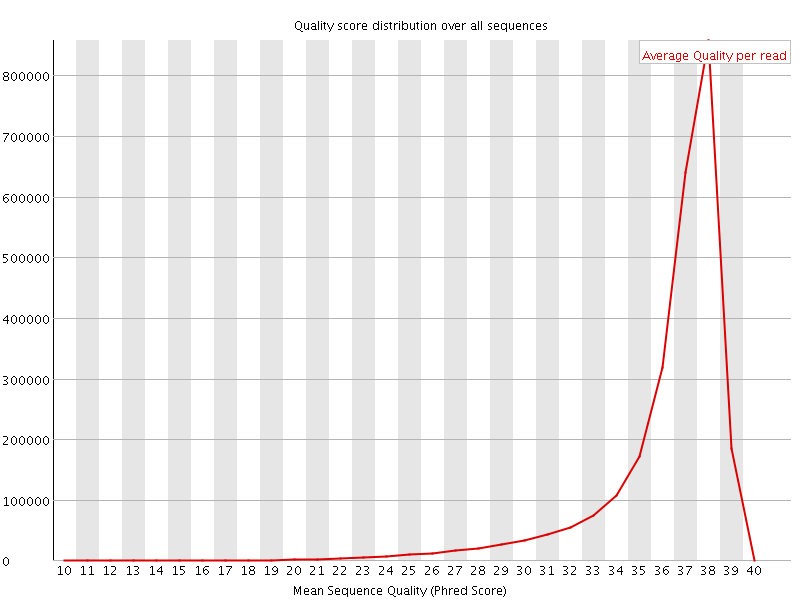
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
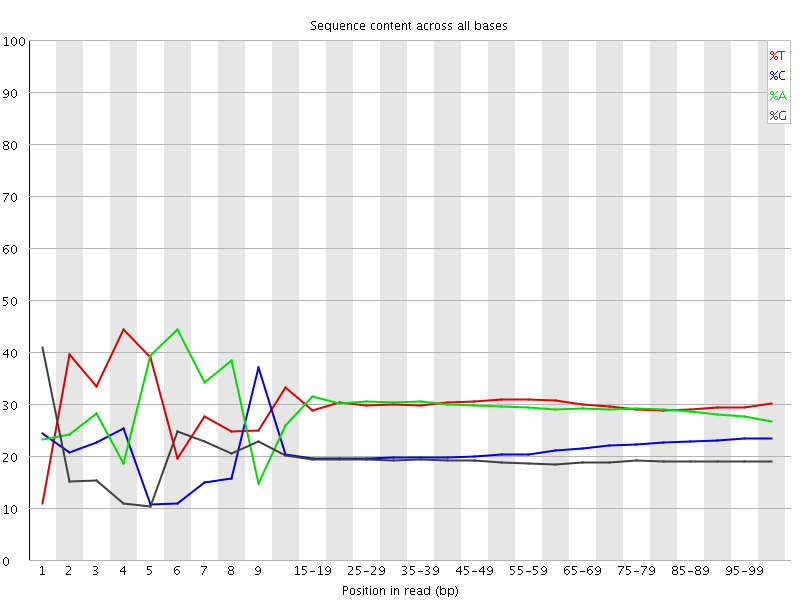
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
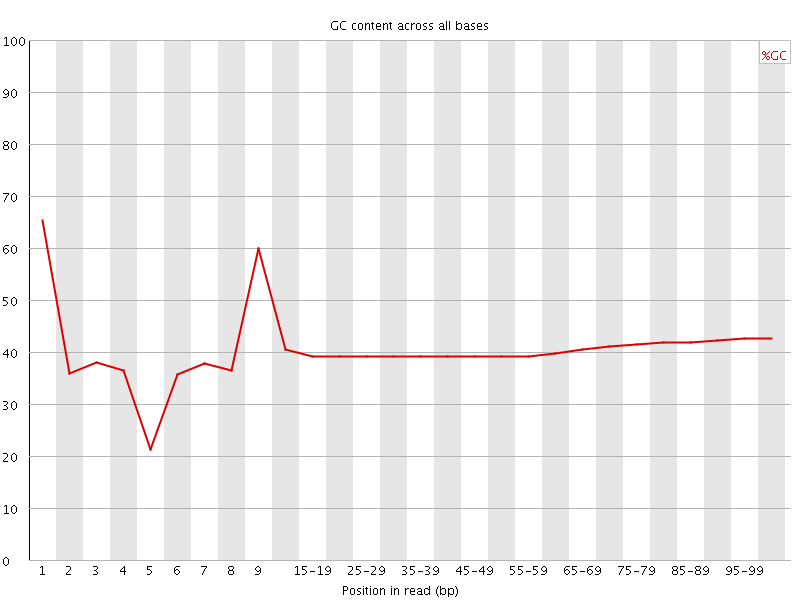
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
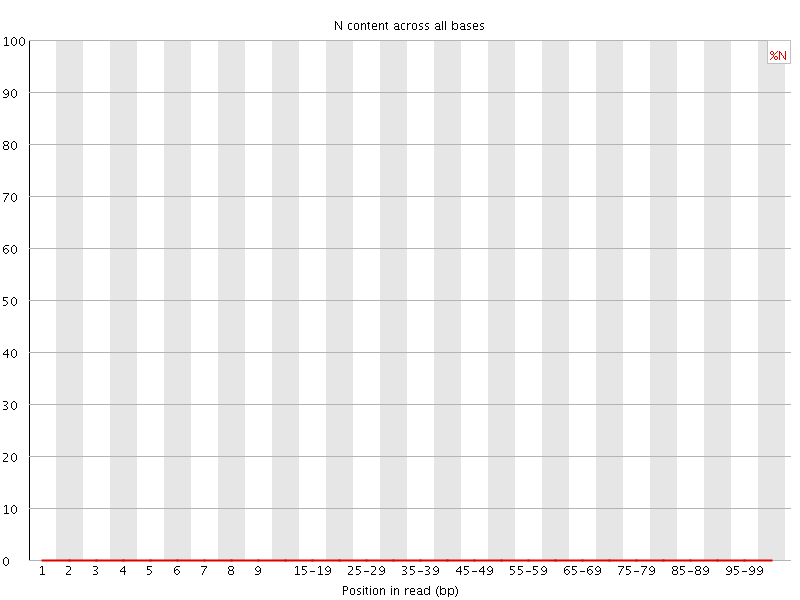
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
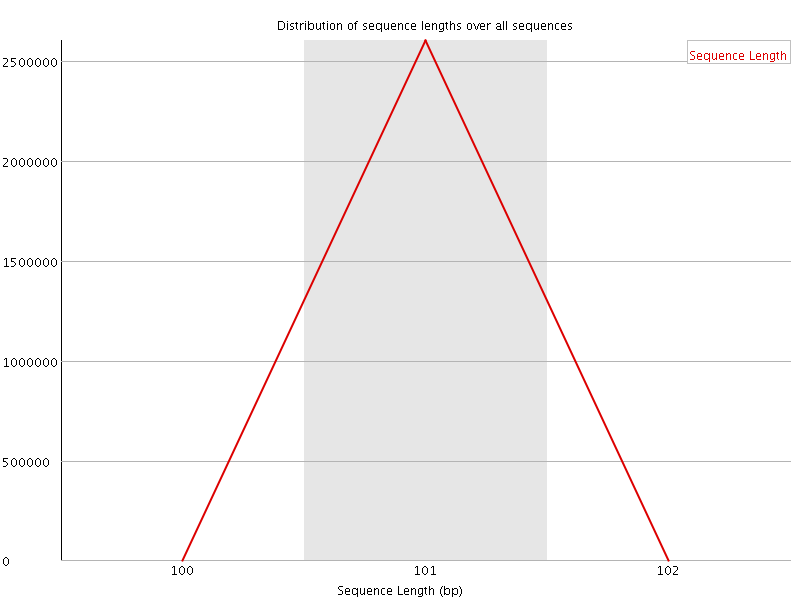
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
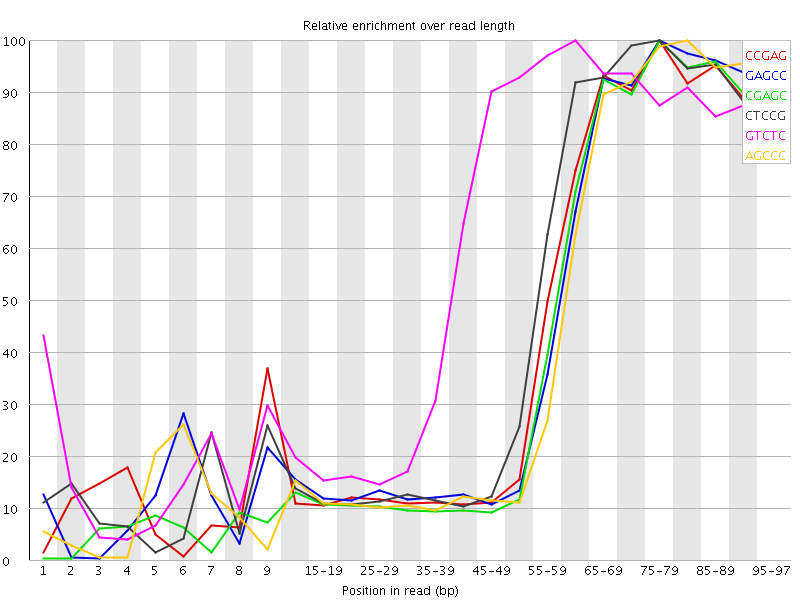
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCGAG | 527285 | 4.237574 | 9.507654 | 75-79 |
| GAGCC | 517720 | 4.160704 | 9.371274 | 75-79 |
| CGAGC | 497765 | 4.000334 | 9.336985 | 75-79 |
| CTCCG | 546495 | 3.9226518 | 8.25227 | 75-79 |
| GTCTC | 755160 | 3.8048165 | 6.253743 | 60-64 |
| AGCCC | 490045 | 3.608843 | 8.405974 | 80-84 |
| CTGTC | 708995 | 3.5722177 | 5.9659896 | 55-59 |
| GCCCA | 483665 | 3.5618584 | 8.556653 | 80-84 |
| TCTCT | 1027870 | 3.3311503 | 4.985947 | 60-64 |
| TCTCC | 697565 | 3.2206204 | 5.973512 | 70-74 |
| CCACG | 430620 | 3.1712186 | 8.349557 | 80-84 |
| CCCAC | 464015 | 3.1312964 | 7.8703146 | 80-84 |
| ATCTC | 938860 | 3.12173 | 7.284907 | 95-97 |
| CTCTT | 956740 | 3.10063 | 4.7402205 | 60-64 |
| TGTCT | 871270 | 3.0814047 | 4.7046194 | 60-64 |
| ACGCT | 577350 | 2.9845054 | 11.065984 | 90-94 |
| TCCGA | 574465 | 2.9695919 | 6.233337 | 75-79 |
| ACACA | 835255 | 2.9234145 | 6.5299025 | 6 |
| GAGAC | 492105 | 2.8481925 | 7.2168183 | 85-89 |
| CGAGA | 491630 | 2.8454432 | 7.0866036 | 85-89 |
| CGCTA | 540250 | 2.7927237 | 10.918605 | 90-94 |
| GACGC | 345155 | 2.7738698 | 8.930575 | 85-89 |
| AGAAG | 649040 | 2.7053442 | 5.3554664 | 5 |
| ACGAG | 465750 | 2.6956553 | 6.7318797 | 85-89 |
| TACAC | 771120 | 2.6306005 | 7.967276 | 5 |
| CACGA | 451405 | 2.3940763 | 6.1211667 | 80-84 |
| AGACG | 408760 | 2.3658102 | 6.726267 | 85-89 |
| GCTAC | 424230 | 2.1929796 | 6.5338182 | 90-94 |
| CTTTG | 552165 | 1.9528319 | 8.882313 | 9 |
| AGAGA | 456120 | 1.9012104 | 5.4464083 | 8 |
| TATAC | 790330 | 1.8925291 | 5.651777 | 5 |
| GAGAG | 289535 | 1.8287421 | 6.3298326 | 7 |
| TGGAG | 279400 | 1.7200433 | 5.035335 | 5 |
| CTACG | 330140 | 1.7065984 | 5.8876553 | 90-94 |
| TCTCG | 331165 | 1.6685498 | 5.5905285 | 90-94 |
| GCTTT | 465020 | 1.6446277 | 5.1399107 | 1 |
| TACGC | 313365 | 1.6198832 | 5.8198886 | 95-97 |
| ATGGA | 374575 | 1.5217786 | 5.0445514 | 4 |
| CTCGT | 285195 | 1.4369334 | 5.548509 | 90-94 |
| GTCTT | 394080 | 1.3937355 | 6.0077105 | 1 |
| GAGTC | 241815 | 1.3641318 | 7.3492174 | 9 |
| AAGAC | 347620 | 1.327749 | 5.8574796 | 5 |
| GACTC | 250865 | 1.2968009 | 5.0903 | 7 |
| ATGAG | 315500 | 1.2817758 | 5.328197 | 7 |
| TATGA | 482530 | 1.2609512 | 5.6596584 | 4 |
| GATTG | 314865 | 1.2468053 | 5.5139017 | 7 |
| TGAGT | 313755 | 1.24241 | 5.6502132 | 8 |
| GCCGT | 154865 | 1.2130724 | 8.013738 | 95-97 |
| AGAAC | 313320 | 1.1967387 | 5.1241364 | 5 |
| TGCCG | 152425 | 1.1939597 | 8.232755 | 95-97 |
| ATGCC | 230770 | 1.1929233 | 5.7363405 | 95-97 |
| GTGTT | 308940 | 1.1923671 | 5.910454 | 1 |
| GACTT | 328245 | 1.1910568 | 6.3633113 | 7 |
| CCGTC | 162445 | 1.1660037 | 6.6937165 | 95-97 |
| TCATA | 483825 | 1.1585703 | 5.511054 | 2 |
| GTTCT | 320990 | 1.1352395 | 5.073025 | 1 |
| GTGTA | 279550 | 1.1069647 | 9.449318 | 1 |
| TGGAC | 193280 | 1.0903351 | 7.546145 | 5 |
| GTCCA | 194495 | 1.0054064 | 7.219357 | 1 |
| GGACT | 176405 | 0.99513954 | 7.094853 | 6 |
| GAGTA | 237165 | 0.9635257 | 5.366519 | 1 |
| AGACT | 252585 | 0.9403301 | 5.402314 | 6 |
| GGATA | 218285 | 0.8868223 | 5.1813307 | 1 |
| GTATA | 282310 | 0.7377347 | 7.301634 | 1 |