![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-157_CAGAGAGG-GTAAGGAG_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
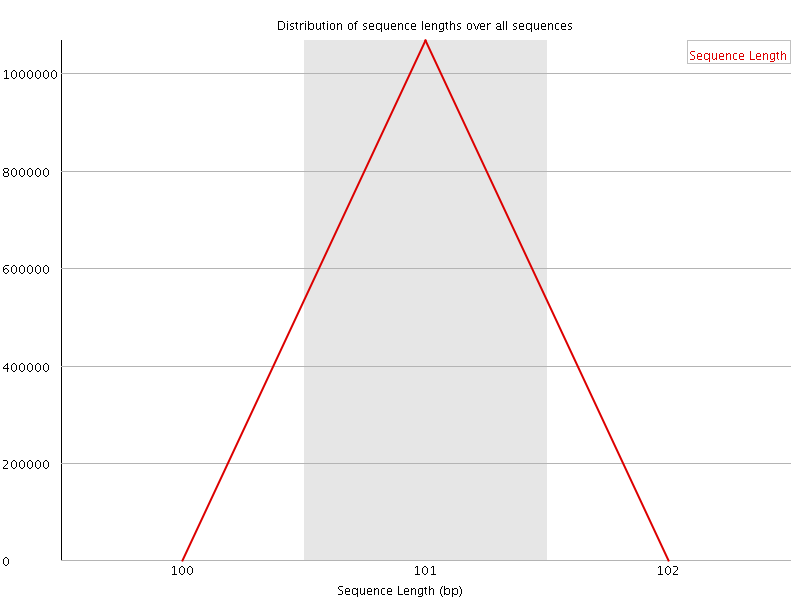
| Total Sequences | 1066943 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
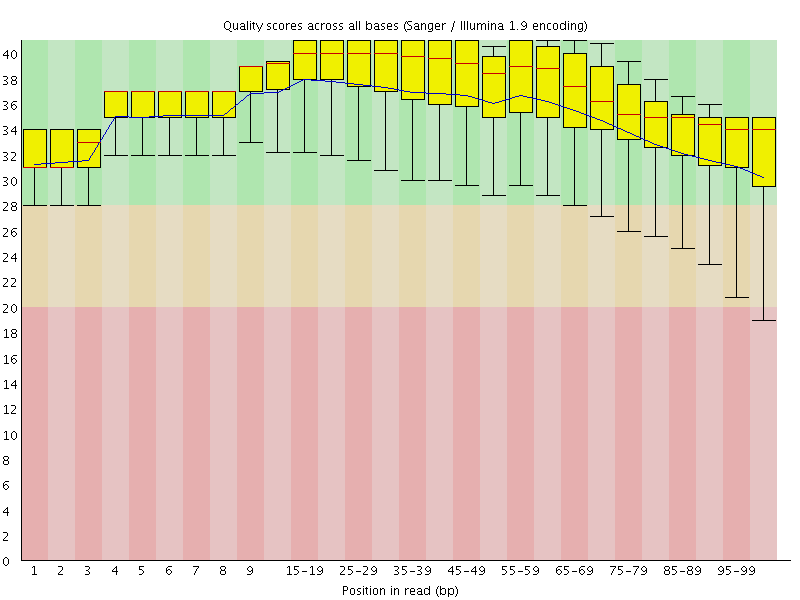
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
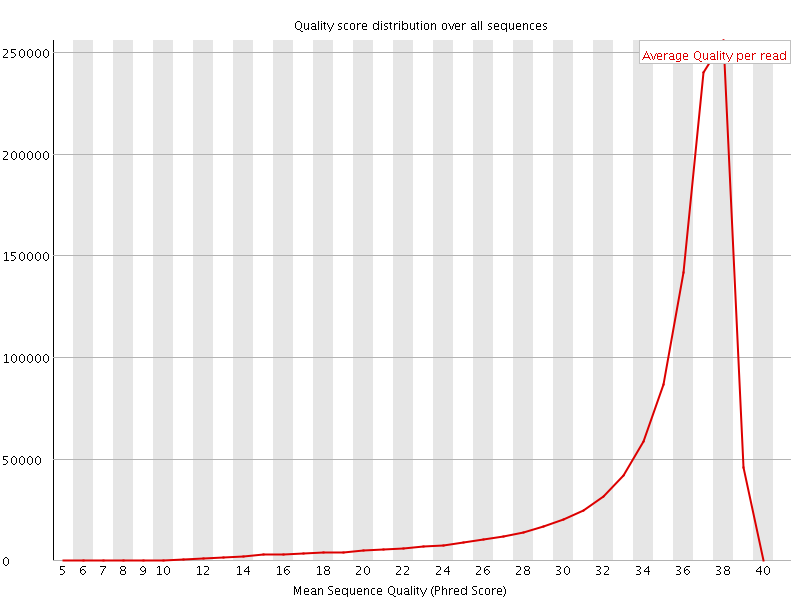
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
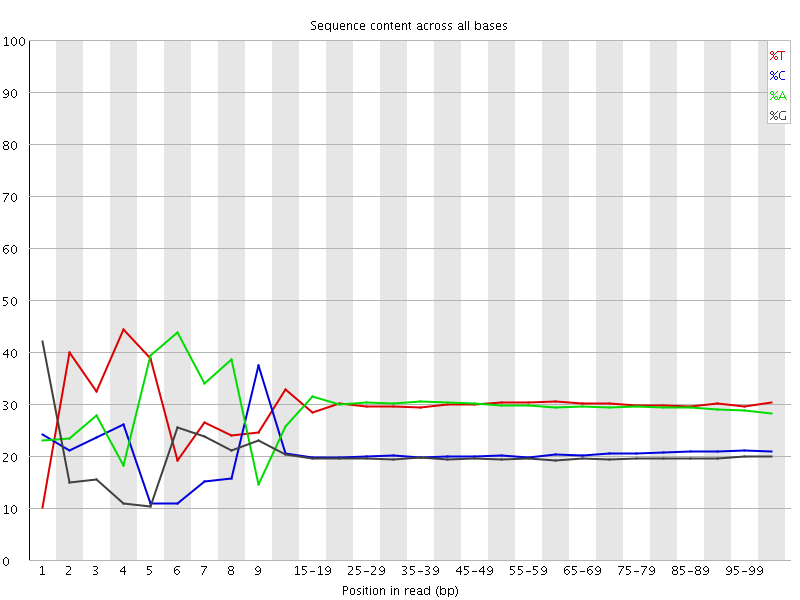
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
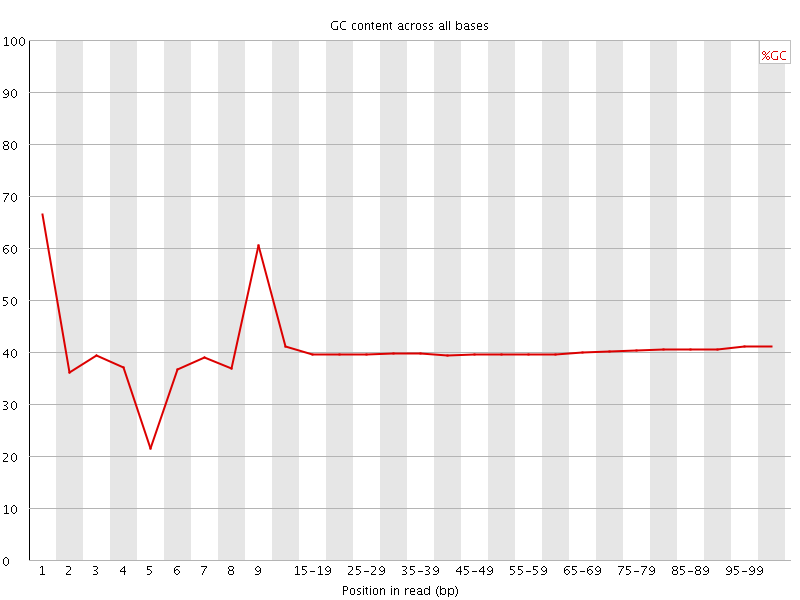
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
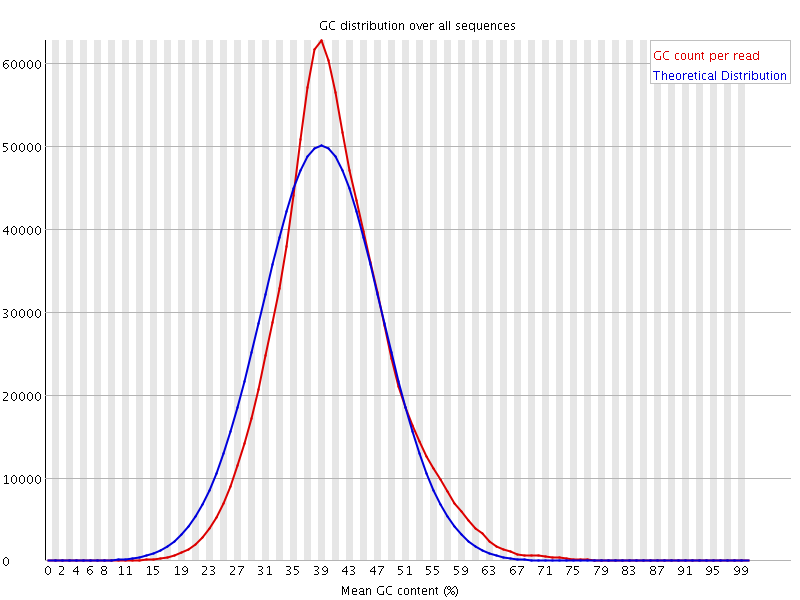
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
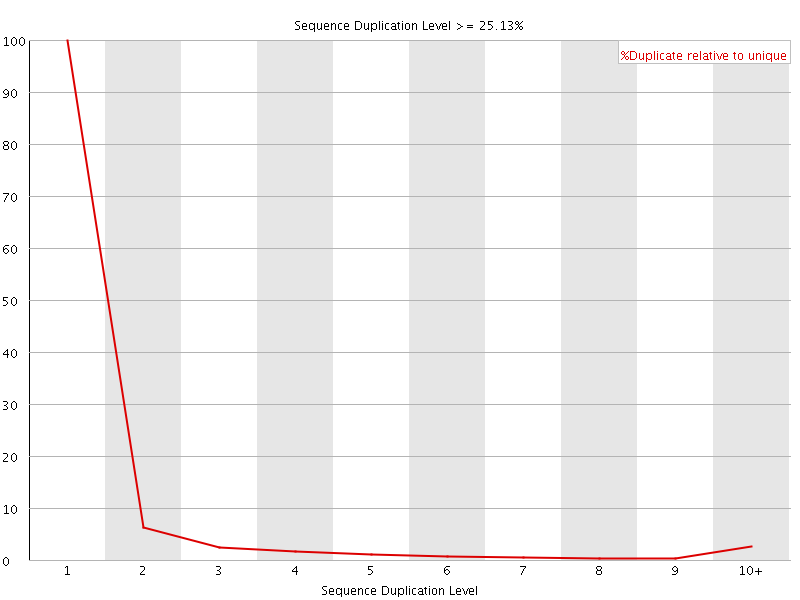
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
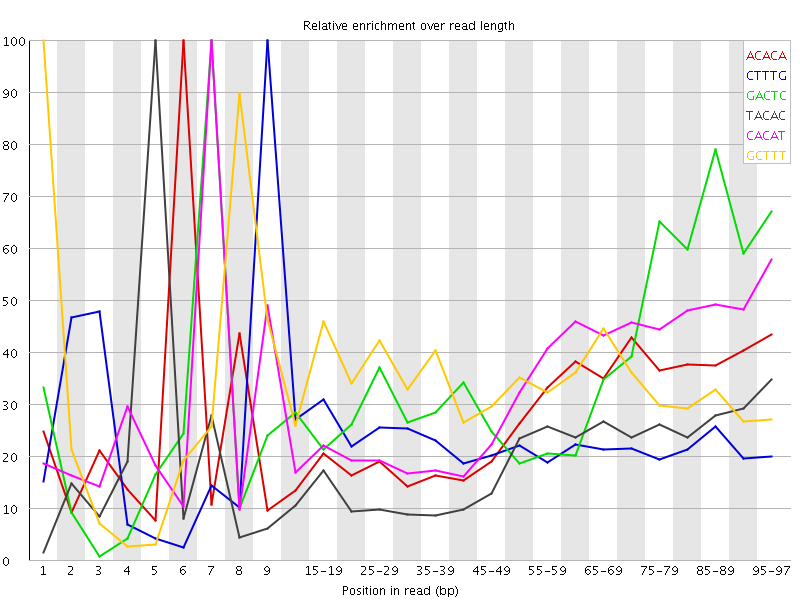
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| ACACA | 248840 | 2.2022061 | 7.943925 | 6 |
| CTTTG | 244210 | 2.1472533 | 9.282809 | 9 |
| GACTC | 157705 | 2.0610356 | 5.6279745 | 7 |
| TACAC | 215190 | 1.8818189 | 9.6463585 | 5 |
| CACAT | 209745 | 1.8342029 | 5.5767426 | 7 |
| GCTTT | 205225 | 1.8044717 | 5.2889175 | 1 |
| GAGAG | 128530 | 1.8025308 | 6.0393505 | 7 |
| GCCAA | 124460 | 1.6460835 | 5.260914 | 1 |
| TGGCT | 122540 | 1.629536 | 5.3524814 | 6 |
| ATACA | 259925 | 1.5662013 | 5.1399193 | 6 |
| TTGAG | 166190 | 1.5227715 | 5.5677795 | 9 |
| CCATG | 110155 | 1.439608 | 5.380825 | 9 |
| GAGTC | 105900 | 1.4251615 | 7.5379148 | 9 |
| AAGAC | 154920 | 1.4118004 | 5.8661604 | 5 |
| GTGTA | 151005 | 1.383634 | 10.832051 | 1 |
| GTCTT | 152285 | 1.3389888 | 6.811614 | 1 |
| GACTT | 146860 | 1.3067886 | 6.6238313 | 7 |
| ATGAG | 140210 | 1.3001422 | 5.2658424 | 7 |
| TATGA | 211480 | 1.2966264 | 6.290325 | 4 |
| TATAC | 216140 | 1.2869234 | 6.1433067 | 5 |
| TCATA | 214645 | 1.2780219 | 6.5331283 | 2 |
| TGAGT | 139295 | 1.2763371 | 5.514509 | 8 |
| ACTCA | 145465 | 1.2720795 | 5.229065 | 8 |
| GATTG | 137120 | 1.256408 | 6.216511 | 7 |
| GTGTT | 137310 | 1.2432258 | 6.1182084 | 1 |
| AACAC | 137710 | 1.2187182 | 5.1815805 | 5 |
| GTTCT | 137665 | 1.2104403 | 5.143899 | 1 |
| TGGAC | 85265 | 1.1474636 | 7.663963 | 5 |
| ACACC | 89160 | 1.1451545 | 5.269329 | 6 |
| ACATG | 124570 | 1.1217531 | 5.7645173 | 8 |
| GTCCA | 84905 | 1.1096176 | 8.203508 | 1 |
| GGACT | 78205 | 1.0524528 | 7.2897434 | 6 |
| ACACT | 119655 | 1.0463731 | 5.4027658 | 6 |
| AGACT | 112535 | 1.0133779 | 5.5678167 | 6 |
| GAGTA | 105710 | 0.98022985 | 5.555257 | 1 |
| GTATG | 105610 | 0.96768713 | 5.55583 | 3 |
| CATAC | 106705 | 0.9331265 | 5.476348 | 3 |
| ATACT | 131455 | 0.78269875 | 5.2262125 | 6 |
| GTATA | 125315 | 0.7683315 | 7.7150884 | 1 |