![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-154_CAGAGAGG-CTCTCTAT_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1786736 |
| Filtered Sequences | 0 |
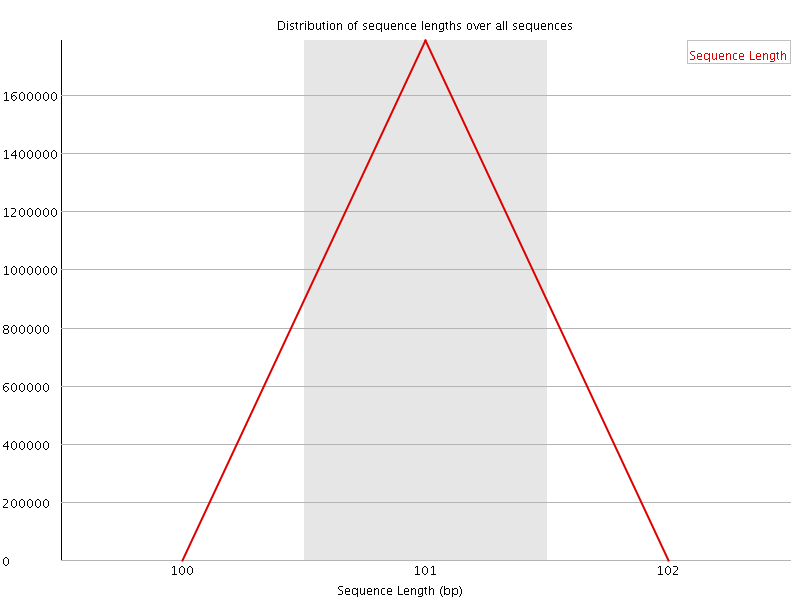
| Sequence length | 101 |
| %GC | 40 |
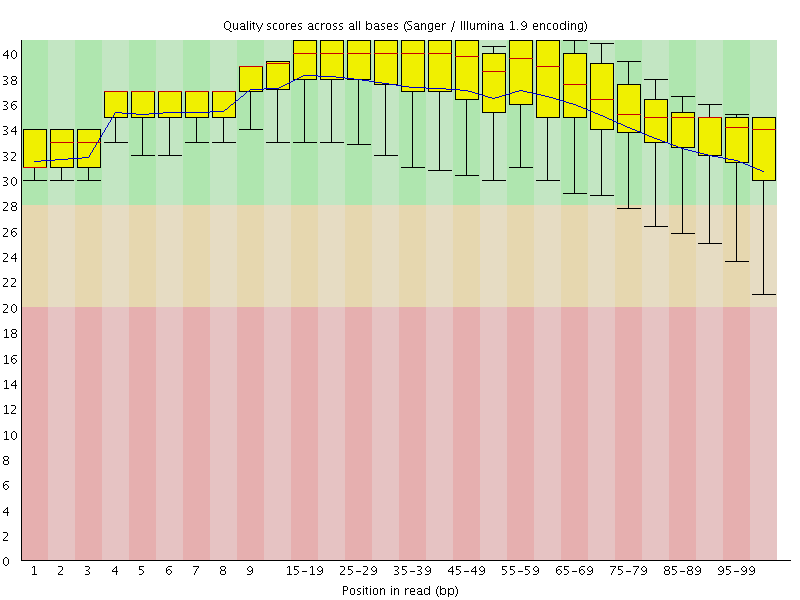
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
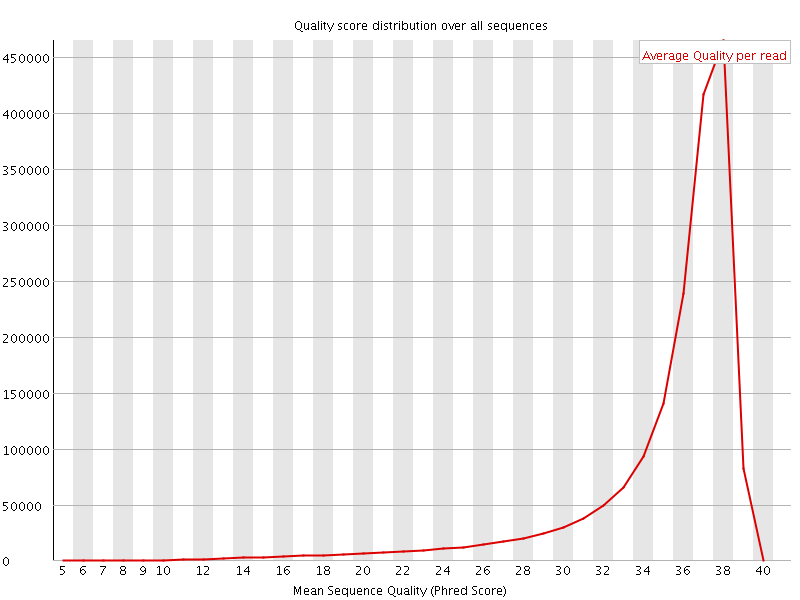
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

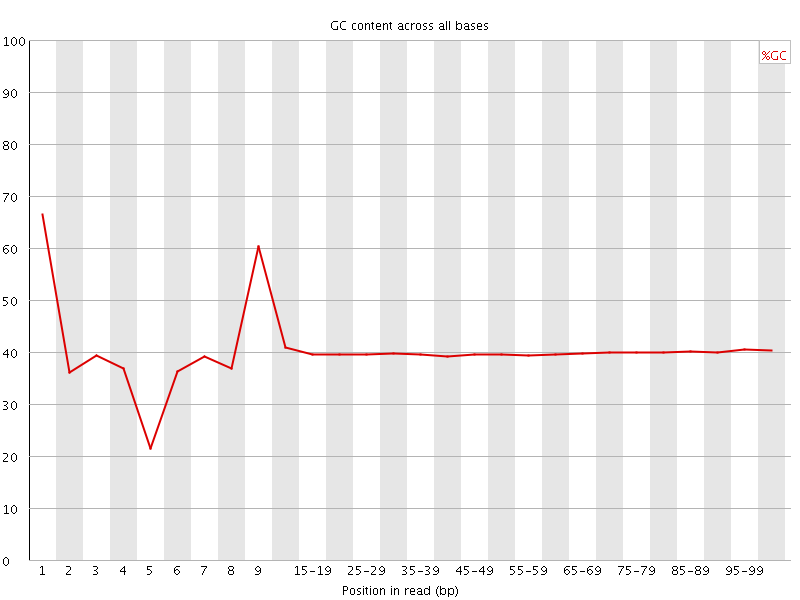
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
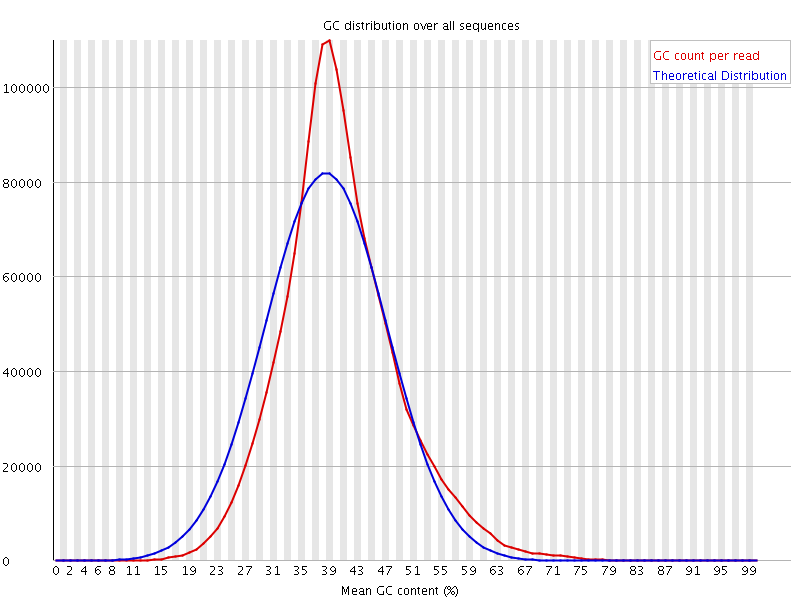
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
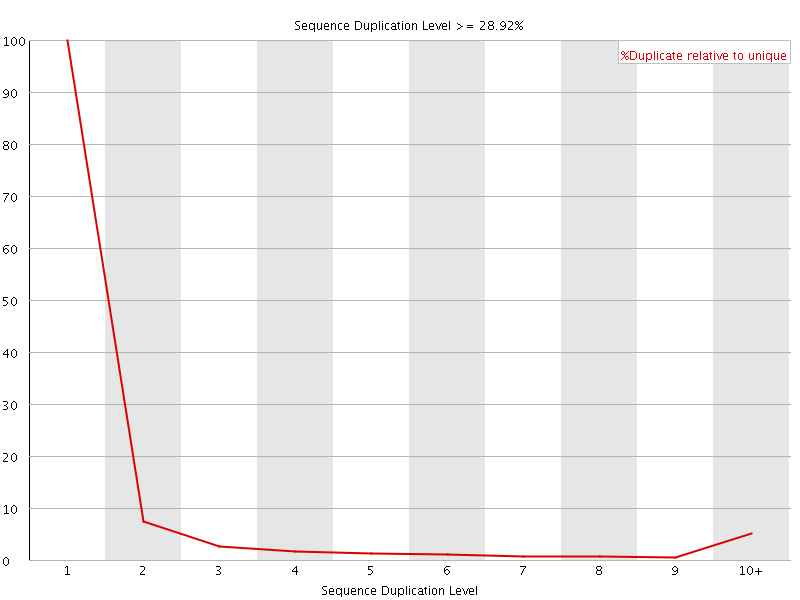
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATATGGACTTTGGCTACACCATGAAAGCTTTGAGAAGCAAGAAGAAGGT | 1824 | 0.10208559070842027 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
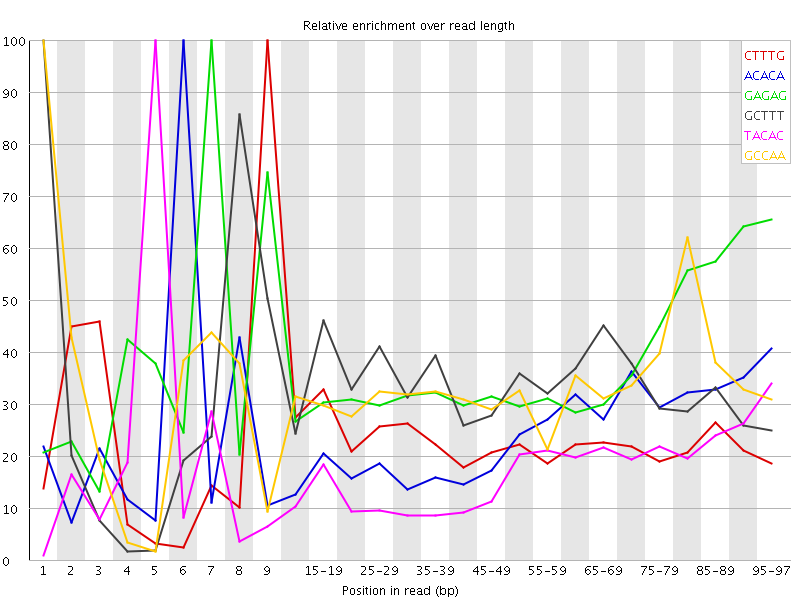
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CTTTG | 441055 | 2.3344233 | 10.041283 | 9 |
| ACACA | 397005 | 2.1119294 | 8.551838 | 6 |
| GAGAG | 252430 | 2.0735977 | 5.485513 | 7 |
| GCTTT | 367165 | 1.9433368 | 5.7667527 | 1 |
| TACAC | 342210 | 1.8088115 | 10.292133 | 5 |
| GCCAA | 222720 | 1.7784375 | 5.2835827 | 1 |
| CACAT | 326880 | 1.7277819 | 6.0878506 | 7 |
| TGGCT | 214585 | 1.7157937 | 5.5565395 | 6 |
| GACTC | 205225 | 1.6282712 | 6.179336 | 7 |
| TTGAG | 295455 | 1.5962951 | 6.060395 | 9 |
| GGCTT | 197370 | 1.5781447 | 5.046354 | 1 |
| GAGTC | 195460 | 1.5729194 | 9.093016 | 9 |
| ATACA | 419880 | 1.4996223 | 5.3432364 | 6 |
| CCATG | 187020 | 1.4838313 | 5.9408317 | 9 |
| ATGGA | 270990 | 1.4735266 | 5.0934777 | 4 |
| AAGAC | 273090 | 1.4734702 | 6.4360814 | 5 |
| CATGA | 271715 | 1.456687 | 5.568979 | 9 |
| TCCAT | 272615 | 1.4317508 | 5.324624 | 2 |
| GTCTT | 269090 | 1.424244 | 6.9529686 | 1 |
| TGTGT | 260930 | 1.4007574 | 5.31622 | 8 |
| GACTT | 261195 | 1.3913442 | 7.561226 | 7 |
| TATGA | 384805 | 1.3850536 | 6.919543 | 4 |
| TCATA | 385600 | 1.3683931 | 7.1716337 | 2 |
| GTGTA | 252935 | 1.3665665 | 11.30148 | 1 |
| TGAGT | 252425 | 1.3638111 | 6.2831774 | 8 |
| ACTCA | 255870 | 1.352446 | 5.521468 | 8 |
| ATGAG | 242845 | 1.3204862 | 5.75386 | 7 |
| GTGTT | 243605 | 1.3077513 | 6.205799 | 1 |
| AACAC | 242310 | 1.2890056 | 5.442363 | 5 |
| GATTG | 237275 | 1.281958 | 6.6472507 | 7 |
| GTTCT | 233635 | 1.2365873 | 5.317429 | 1 |
| ACACC | 153665 | 1.2097685 | 5.3335485 | 6 |
| ACCAT | 226320 | 1.1962543 | 5.0011067 | 8 |
| TATAC | 335795 | 1.1916481 | 6.2678976 | 5 |
| TGGAC | 147730 | 1.1888233 | 8.494444 | 5 |
| GTCCA | 149260 | 1.1842407 | 8.863908 | 1 |
| ACATG | 216530 | 1.1608355 | 6.2138224 | 8 |
| ATGTG | 210600 | 1.1378373 | 5.4053125 | 7 |
| ACACT | 212140 | 1.1213033 | 5.549463 | 6 |
| GGACT | 137900 | 1.1097187 | 8.101557 | 6 |
| AGACT | 194390 | 1.0421412 | 5.971812 | 6 |
| GTATG | 192535 | 1.040235 | 6.232395 | 3 |
| TGTAT | 287965 | 1.0298707 | 5.2757525 | 2 |
| CATAC | 188115 | 0.994315 | 5.981866 | 3 |
| GAGTA | 182560 | 0.99268234 | 5.1120276 | 1 |
| ATACT | 226020 | 0.8020855 | 5.305665 | 6 |
| GTATA | 217245 | 0.781944 | 7.588402 | 1 |