![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-151_CTCTCTAC-AAGGAGTA_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1766755 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 41 |
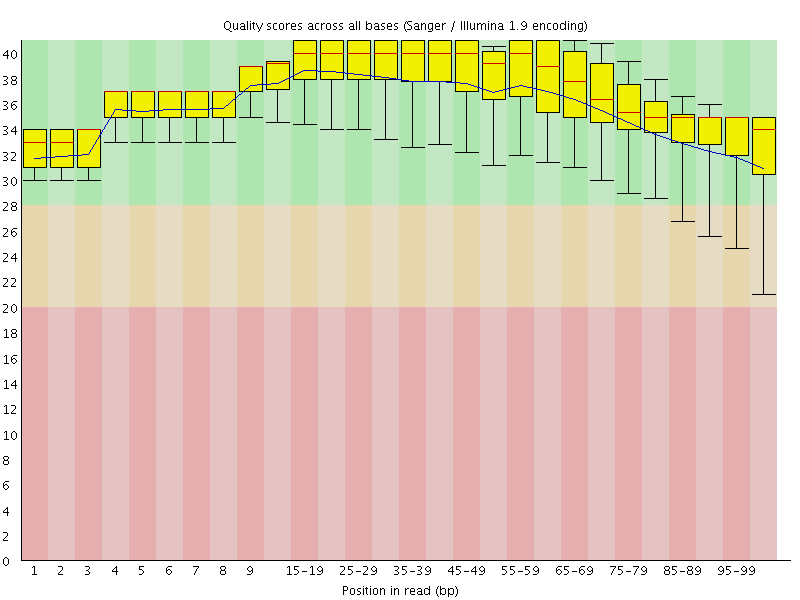
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
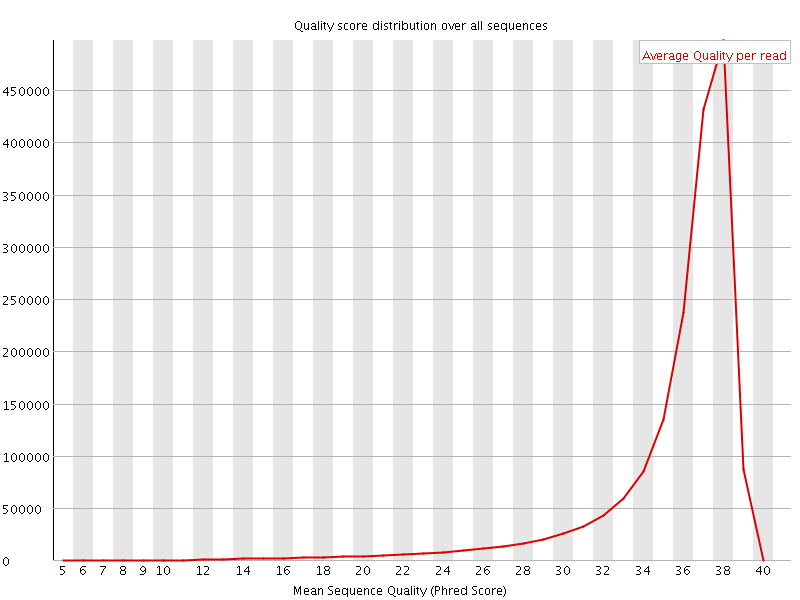
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
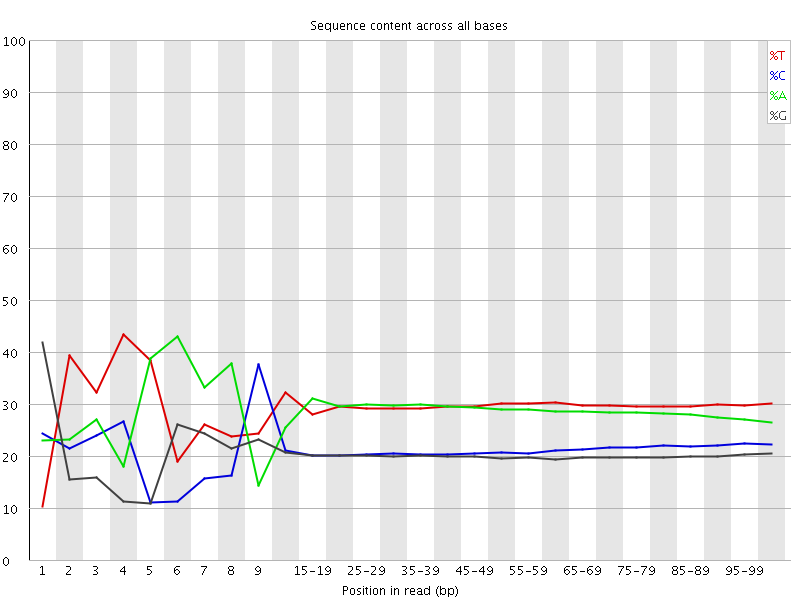
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
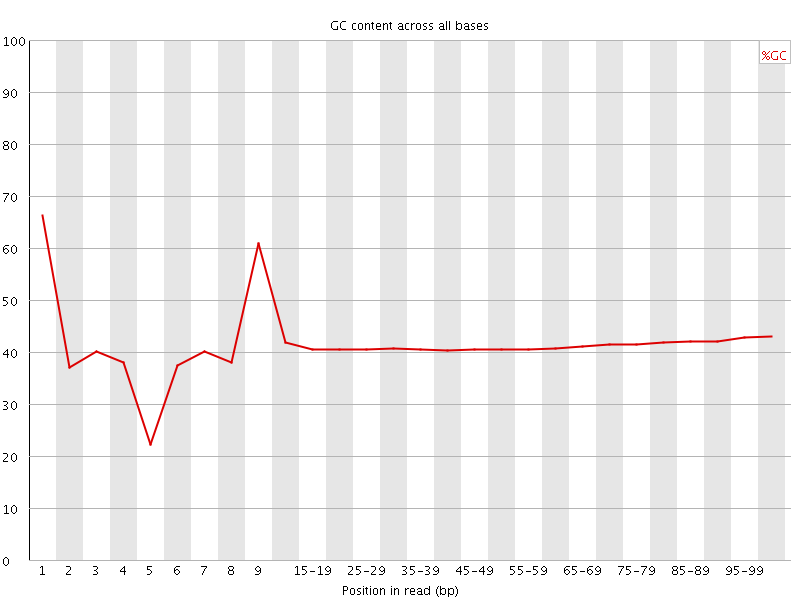
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
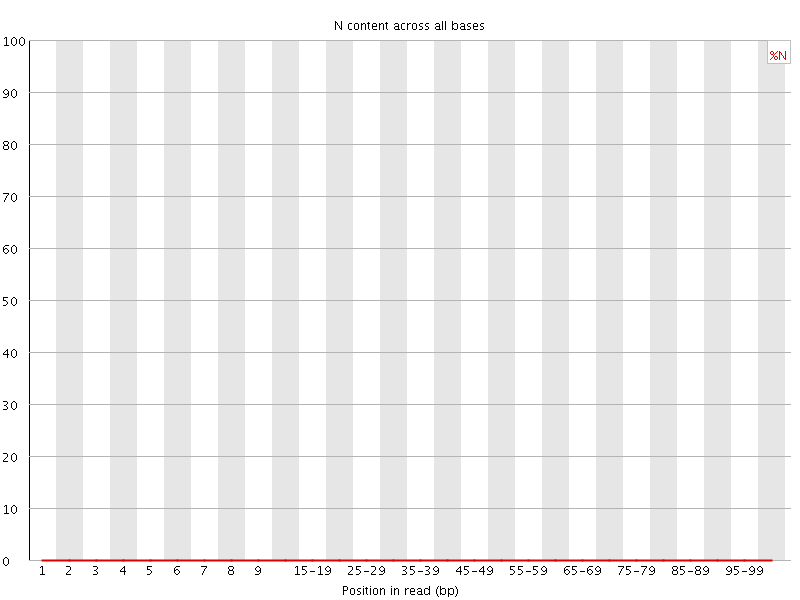
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
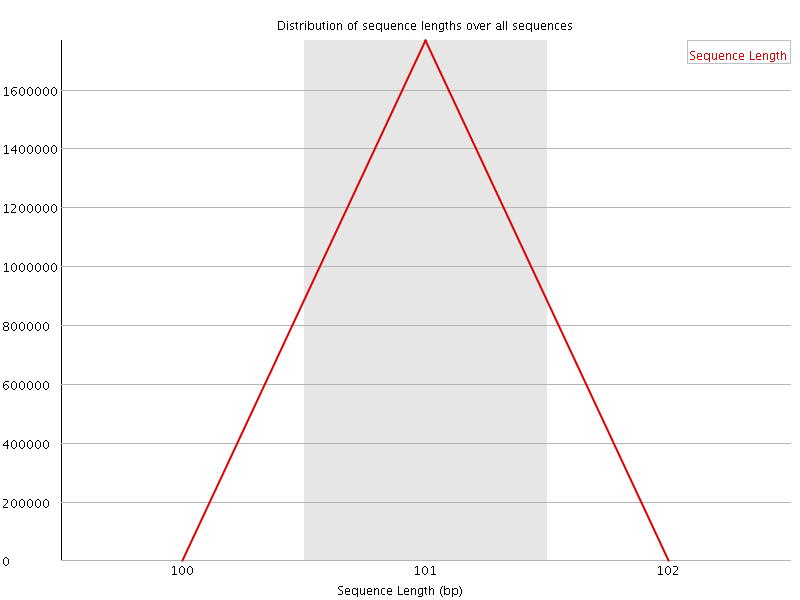
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
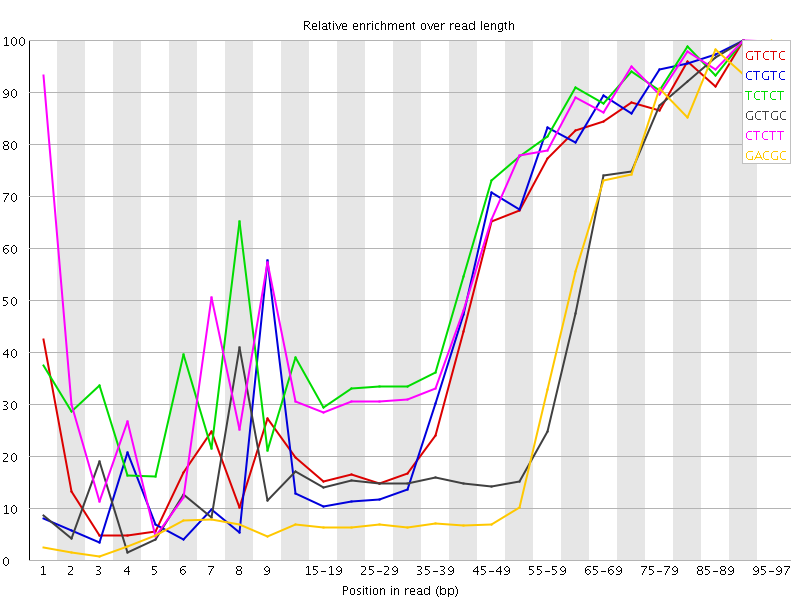
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GTCTC | 487155 | 3.528747 | 6.343215 | 90-94 |
| CTGTC | 458225 | 3.3191903 | 5.924456 | 90-94 |
| TCTCT | 670120 | 3.2969227 | 5.0618963 | 90-94 |
| GCTGC | 306925 | 3.273277 | 7.787439 | 90-94 |
| CTCTT | 621370 | 3.0570776 | 4.81899 | 90-94 |
| GACGC | 271255 | 2.9906144 | 7.867972 | 95-97 |
| CGCTG | 271055 | 2.890733 | 7.547009 | 95-97 |
| ACACA | 524625 | 2.851685 | 7.3728576 | 6 |
| GCCGA | 255870 | 2.8209932 | 7.8409677 | 90-94 |
| CTGCC | 271160 | 2.7588756 | 7.3622384 | 90-94 |
| CGACG | 245700 | 2.7088678 | 8.033745 | 95-97 |
| AGAAG | 453525 | 2.708582 | 5.3598266 | 5 |
| TGCCG | 241655 | 2.5771894 | 7.5081344 | 90-94 |
| CCGAC | 243310 | 2.5591671 | 7.3263583 | 90-94 |
| TACAC | 483350 | 2.5414538 | 9.032016 | 5 |
| CTGAC | 323475 | 2.4222906 | 5.989288 | 95-97 |
| ATCTG | 451125 | 2.4050784 | 5.0380754 | 95-97 |
| TGACG | 302845 | 2.3771136 | 6.0051246 | 95-97 |
| GACGA | 277985 | 2.2557087 | 6.3182373 | 95-97 |
| CAAAG | 383590 | 2.1855657 | 5.279202 | 4 |
| CCTTG | 289145 | 2.0944455 | 5.065091 | 95-97 |
| ACGCT | 278355 | 2.0844164 | 5.333093 | 95-97 |
| CTCCT | 300010 | 2.0732183 | 5.007865 | 90-94 |
| CTTTG | 375480 | 1.9363639 | 8.165568 | 9 |
| AGAGA | 318420 | 1.901696 | 5.091799 | 8 |
| TATAC | 489890 | 1.8338587 | 5.814695 | 5 |
| GAGAG | 209745 | 1.78401 | 5.9603996 | 7 |
| GGTGG | 151465 | 1.774806 | 5.204093 | 95-97 |
| GCTTT | 320860 | 1.6546868 | 5.1685953 | 1 |
| GTGTA | 266095 | 1.4870074 | 10.233763 | 1 |
| TTGAG | 264930 | 1.480497 | 5.4038663 | 9 |
| AAGAC | 240940 | 1.3727945 | 5.986781 | 5 |
| GAGTC | 169335 | 1.3291569 | 6.25037 | 9 |
| CTCGG | 124370 | 1.3263745 | 5.139967 | 95-97 |
| CCATG | 173000 | 1.2954828 | 5.3891773 | 9 |
| TATGA | 316255 | 1.2409341 | 6.2320123 | 4 |
| GTCTT | 240155 | 1.2384882 | 6.2718625 | 1 |
| GATTG | 215985 | 1.2069798 | 5.9594126 | 7 |
| GACTT | 224405 | 1.1963682 | 6.233492 | 7 |
| TCATA | 316815 | 1.1859683 | 6.186914 | 2 |
| GTGTT | 218595 | 1.181638 | 5.803201 | 1 |
| GTTCT | 221280 | 1.1411492 | 5.2136264 | 1 |
| TGGAC | 138060 | 1.0836709 | 7.2842155 | 5 |
| ACATG | 195530 | 1.0776504 | 5.212076 | 8 |
| GTCCA | 136050 | 1.0187886 | 7.2326717 | 1 |
| GGACT | 125875 | 0.98802745 | 6.6270328 | 6 |
| AGACT | 179015 | 0.98662907 | 5.524535 | 6 |
| GAGTA | 169560 | 0.979562 | 5.490159 | 1 |
| GTATG | 168995 | 0.94438756 | 5.4135747 | 3 |
| GTATA | 188655 | 0.74025214 | 7.029632 | 1 |