![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-128_TCCTGAGC-CTAAGCCT_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1163287 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 39 |
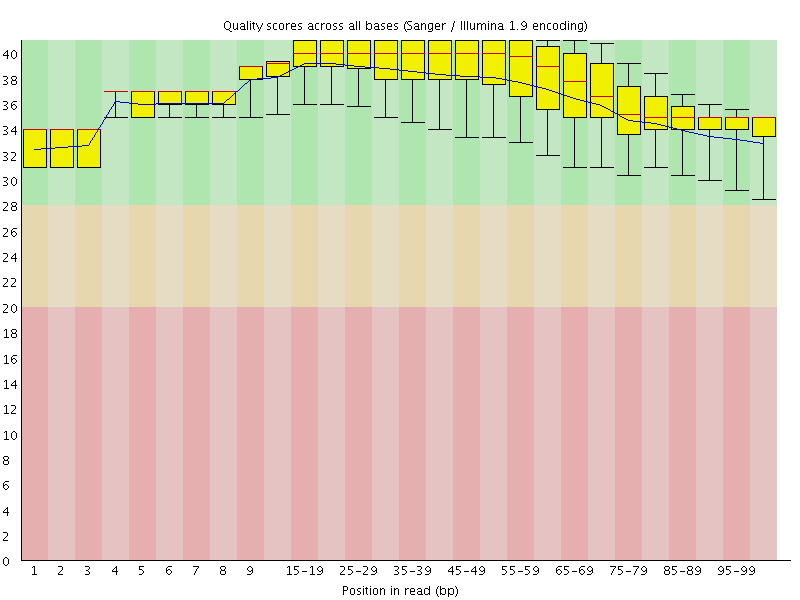
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
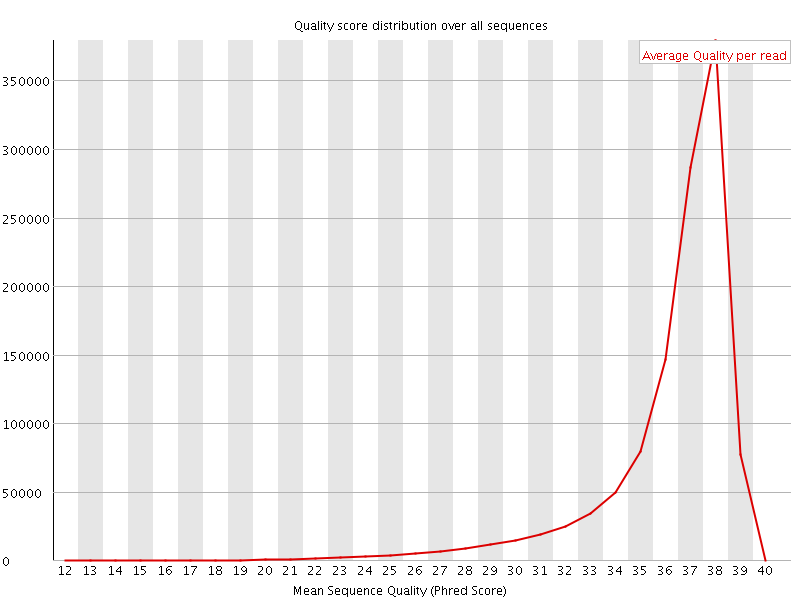
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
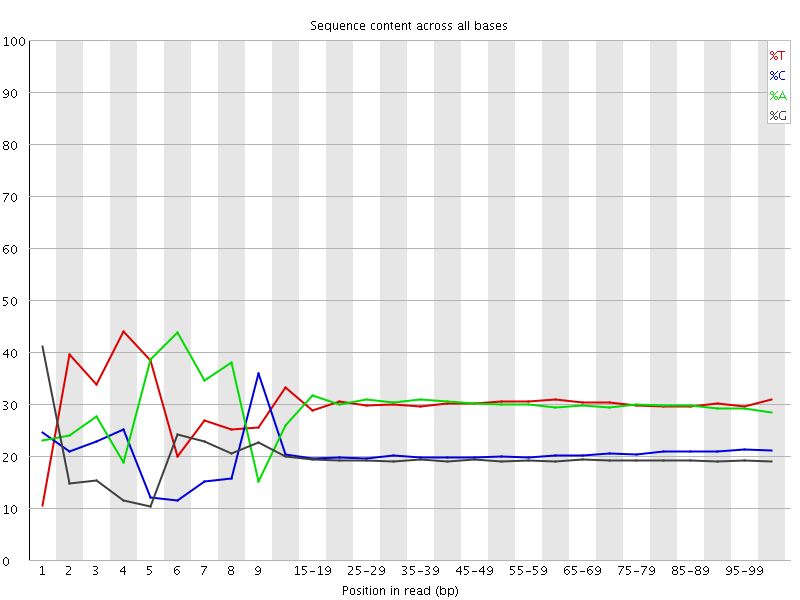
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
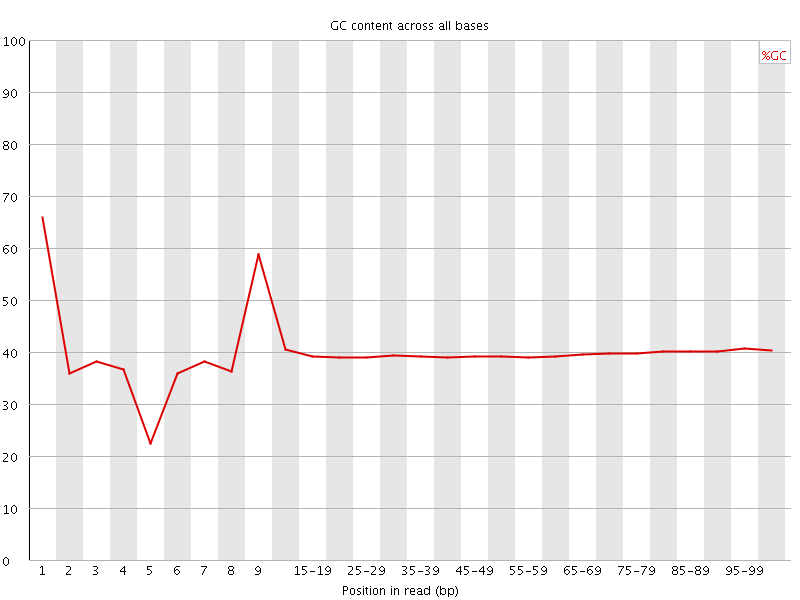
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
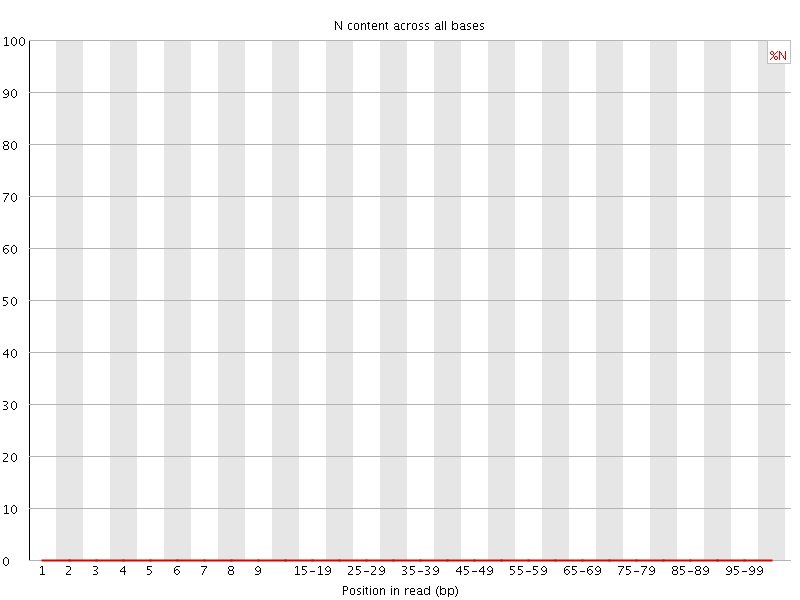
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
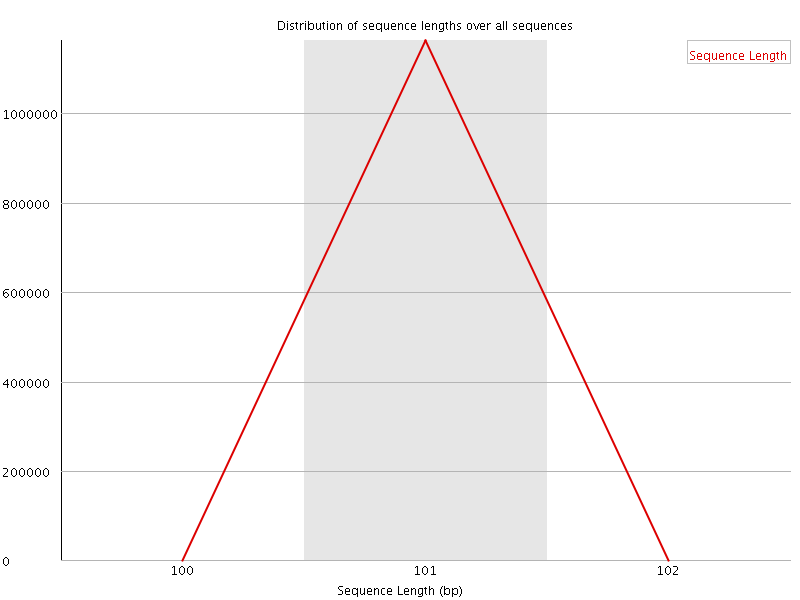
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATATGGACTTTGGCTACACCATGAAAGCTTTGAGAAGCAAGAAGAAGGT | 1252 | 0.10762606304377167 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
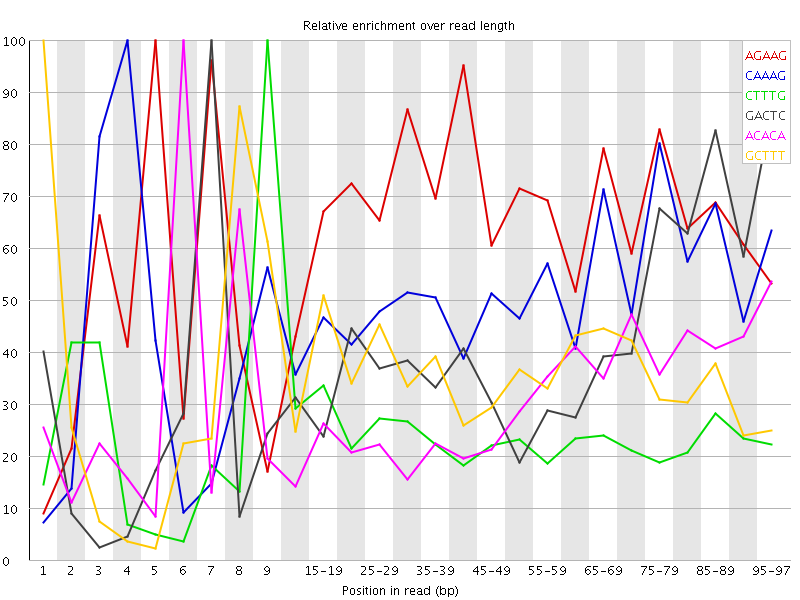
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AGAAG | 345180 | 3.006438 | 4.5396066 | 5 |
| CAAAG | 326660 | 2.7263446 | 5.3333893 | 4 |
| CTTTG | 311700 | 2.5189376 | 10.457606 | 9 |
| GACTC | 195825 | 2.3762195 | 5.718575 | 7 |
| ACACA | 285520 | 2.2834918 | 7.3248377 | 6 |
| GCTTT | 261075 | 2.1098228 | 5.9329066 | 1 |
| GCCAA | 164635 | 2.0193365 | 5.5075502 | 1 |
| TACAC | 253175 | 2.0031605 | 9.41013 | 5 |
| TGGCT | 153705 | 1.925574 | 5.5273466 | 6 |
| GGTTG | 142955 | 1.8689315 | 5.2547455 | 7 |
| CACAT | 230910 | 1.8269962 | 5.0752544 | 7 |
| GAGTC | 140715 | 1.7818893 | 10.247071 | 9 |
| GGCTT | 139325 | 1.7454255 | 5.4855604 | 1 |
| TTGAG | 202330 | 1.7247685 | 5.6251197 | 9 |
| GAGAG | 128070 | 1.7107147 | 5.0321584 | 7 |
| AAGAC | 194830 | 1.6260751 | 5.875631 | 5 |
| CATGA | 192075 | 1.5859429 | 5.0081472 | 9 |
| ATACA | 294380 | 1.5848986 | 5.6644273 | 6 |
| GTCTT | 195235 | 1.5777503 | 6.681377 | 1 |
| ATGGA | 178440 | 1.5375553 | 5.167867 | 4 |
| TGTGT | 182050 | 1.5352994 | 6.7916474 | 8 |
| TCATG | 187740 | 1.5335766 | 5.1763945 | 5 |
| CCATG | 126240 | 1.5318474 | 5.9421415 | 9 |
| AGACT | 184260 | 1.5214154 | 5.8208213 | 6 |
| TGAGT | 178310 | 1.520009 | 8.5389385 | 8 |
| GACTT | 185485 | 1.5151564 | 7.9131107 | 7 |
| TATGA | 267065 | 1.4844444 | 5.9261866 | 4 |
| TCAAG | 177375 | 1.4645667 | 5.352433 | 8 |
| TCATA | 274255 | 1.4607629 | 6.0591836 | 2 |
| GTGTT | 171780 | 1.4486884 | 6.6616907 | 1 |
| ACTCA | 182645 | 1.4451159 | 5.3668036 | 8 |
| GTGTA | 168330 | 1.4349341 | 10.586223 | 1 |
| ATGAG | 166320 | 1.4331216 | 6.663499 | 7 |
| TCCAT | 181335 | 1.4194123 | 5.3822527 | 2 |
| AACAC | 174245 | 1.3935523 | 5.3394923 | 5 |
| CAAGT | 162500 | 1.3417453 | 6.109059 | 9 |
| GATTG | 155275 | 1.3236467 | 6.2450814 | 7 |
| TATAC | 241760 | 1.2876849 | 5.48766 | 5 |
| ACACC | 108560 | 1.2759526 | 5.379474 | 6 |
| GTTCT | 153860 | 1.2433869 | 5.4548244 | 1 |
| GTCCA | 101945 | 1.237042 | 8.779065 | 1 |
| GTATG | 145030 | 1.2363129 | 5.174614 | 3 |
| TGGAC | 97500 | 1.234653 | 8.128892 | 5 |
| ACATG | 147380 | 1.2169011 | 5.4565187 | 8 |
| ATGTG | 142175 | 1.2119753 | 6.9229064 | 7 |
| ACACT | 152900 | 1.2097688 | 5.493397 | 6 |
| GGACT | 94985 | 1.2028054 | 8.853372 | 6 |
| GTATA | 152105 | 0.84545493 | 7.2395897 | 1 |