![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-125_TCCTGAGC-GTAAGGAG_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 843328 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 39 |
![[OK]](Icons/tick.png) Per base sequence quality
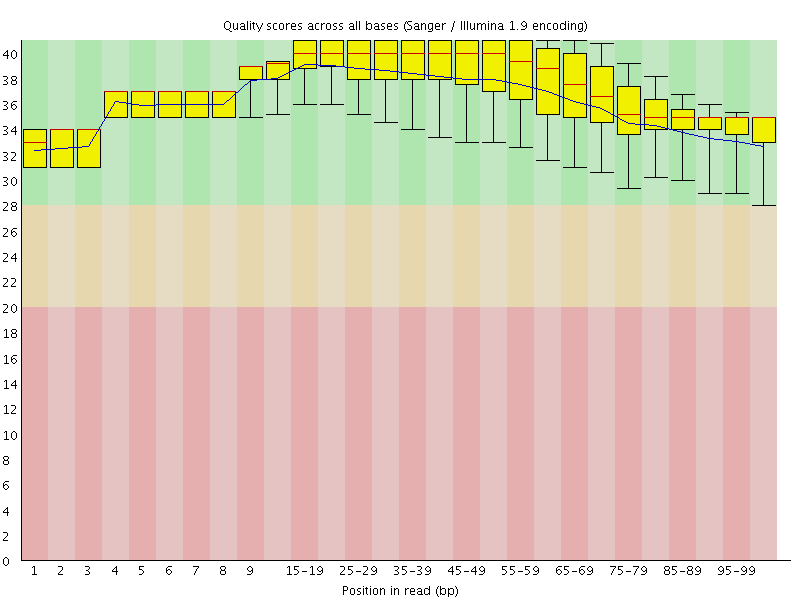
Per base sequence quality
![[OK]](Icons/tick.png) Per sequence quality scores
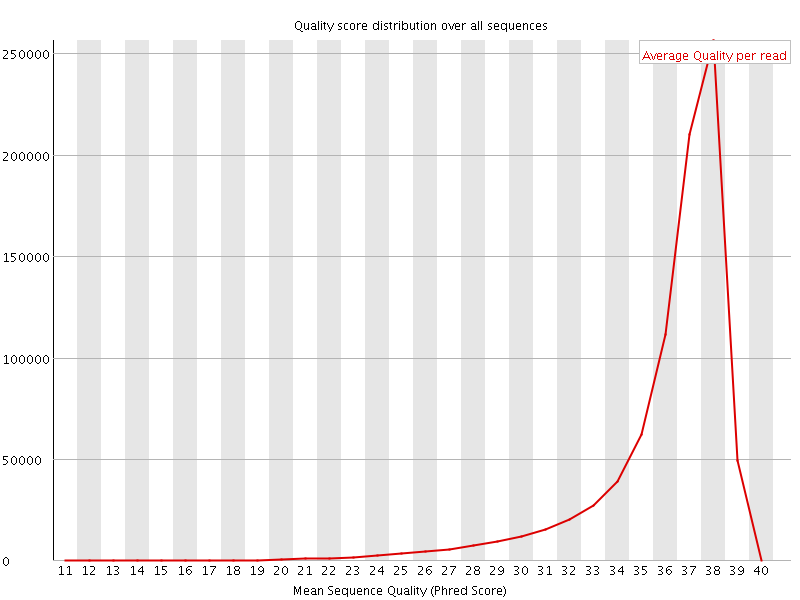
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
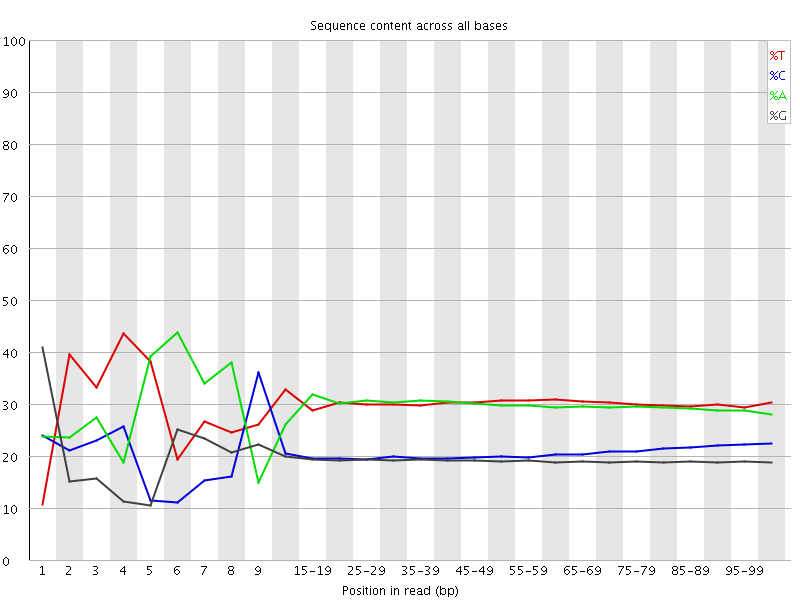
Per base sequence content
![[FAIL]](Icons/error.png) Per base GC content
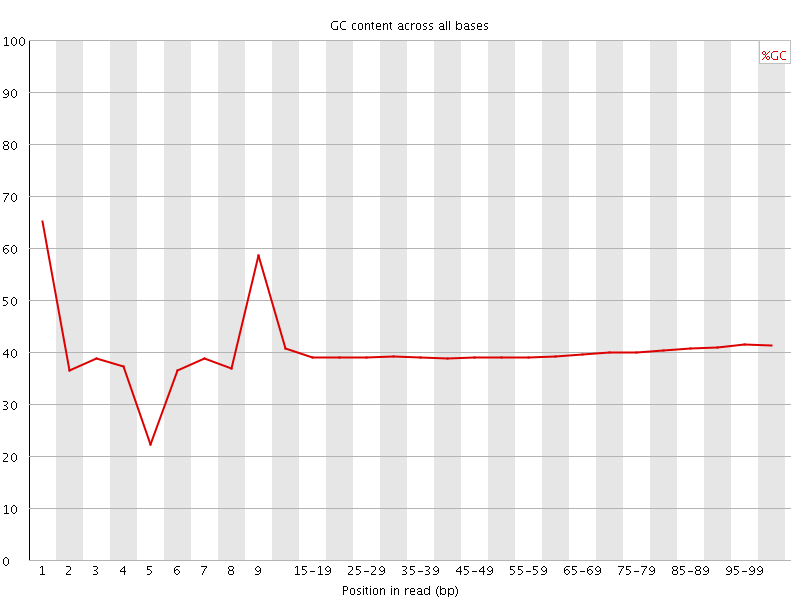
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
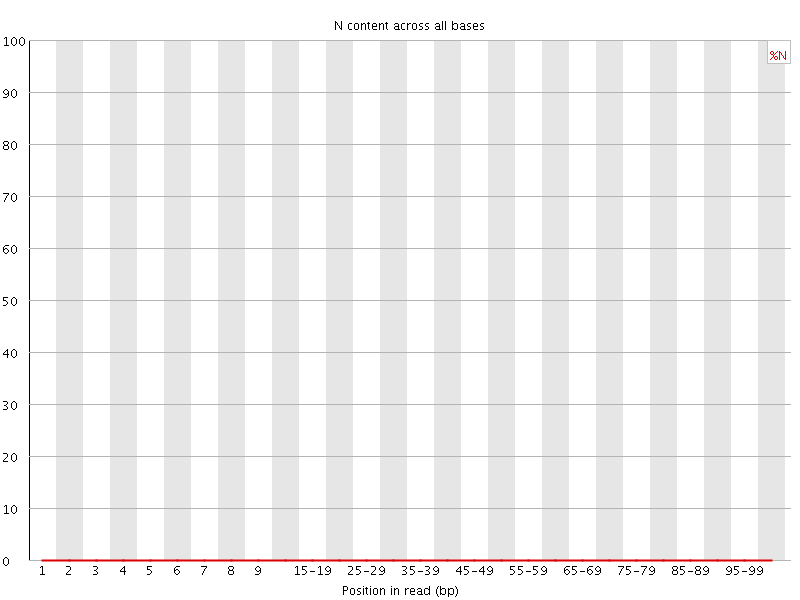
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
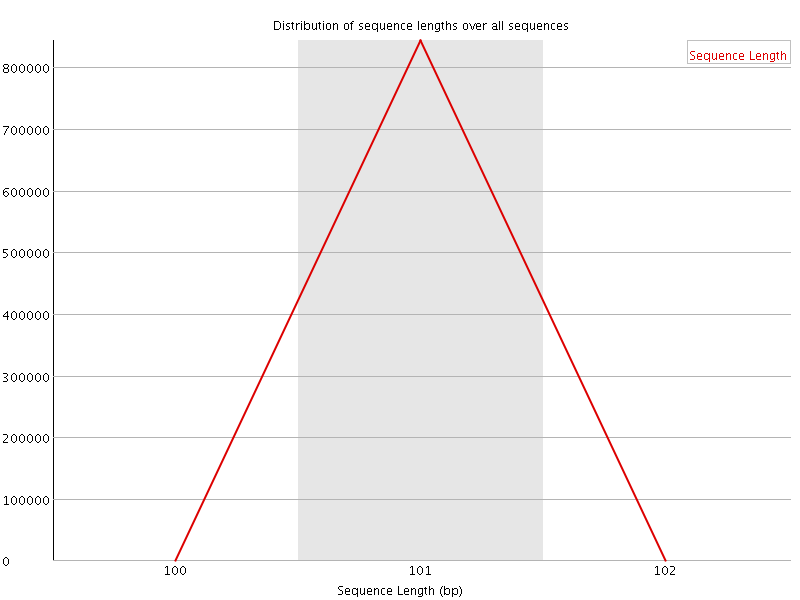
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
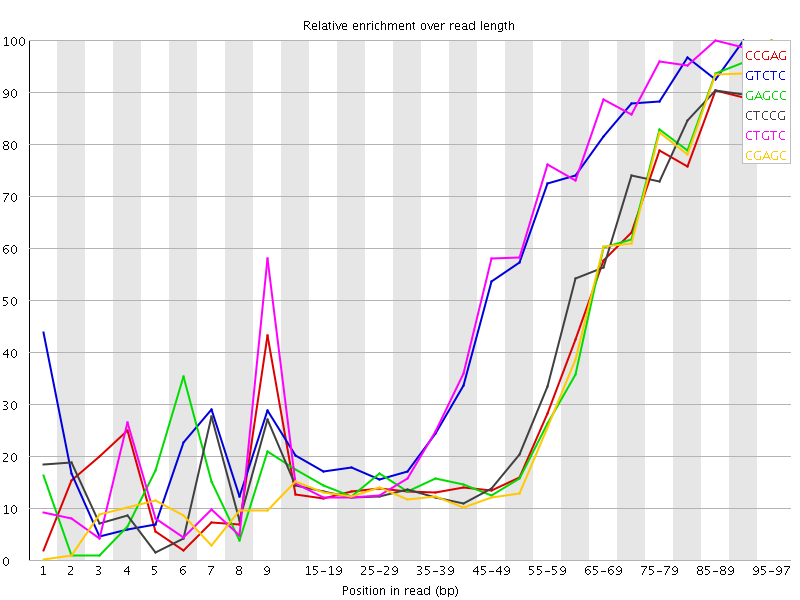
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CCGAG | 121710 | 3.1433234 | 8.302067 | 95-97 |
| GTCTC | 190480 | 3.0927446 | 5.75016 | 90-94 |
| GAGCC | 117295 | 3.0293 | 7.8011875 | 95-97 |
| CTCCG | 125485 | 2.998703 | 7.6045933 | 95-97 |
| CTGTC | 178855 | 2.9039943 | 5.397633 | 85-89 |
| CGAGC | 111850 | 2.888676 | 7.767797 | 95-97 |
| AGAAG | 228990 | 2.8304858 | 5.0279655 | 5 |
| GCCCA | 110050 | 2.677747 | 7.1212 | 90-94 |
| AGCCC | 109930 | 2.674827 | 7.0763683 | 90-94 |
| TCTCC | 171405 | 2.6220126 | 5.567604 | 95-97 |
| ACACA | 227725 | 2.4985528 | 6.6918206 | 6 |
| CCCAC | 103875 | 2.3812616 | 6.7047396 | 90-94 |
| CAAAG | 201045 | 2.3412855 | 5.3411913 | 4 |
| GACTC | 141490 | 2.339151 | 5.6995416 | 85-89 |
| CATCT | 218635 | 2.3137789 | 5.9178905 | 95-97 |
| ATCTC | 218525 | 2.3126147 | 5.789613 | 95-97 |
| CCACG | 93350 | 2.2714007 | 6.717712 | 90-94 |
| TCCGA | 137290 | 2.2697153 | 5.4720626 | 95-97 |
| TACAC | 210210 | 2.265131 | 8.562583 | 5 |
| CTTTG | 196900 | 2.172168 | 8.809011 | 9 |
| GAGAC | 113290 | 2.0241618 | 5.558697 | 95-97 |
| CGAGA | 112555 | 2.0110297 | 5.500945 | 95-97 |
| ATACA | 250995 | 1.8710951 | 5.504483 | 6 |
| ACGAG | 104625 | 1.8693435 | 5.3247986 | 95-97 |
| GCTTT | 166710 | 1.8391169 | 5.4830775 | 1 |
| GCCAA | 100265 | 1.6877948 | 5.0362616 | 1 |
| TGGCT | 97910 | 1.6873497 | 5.096755 | 6 |
| GAGAG | 88025 | 1.6693331 | 5.6729374 | 7 |
| AGACT | 139675 | 1.5975041 | 5.567278 | 6 |
| TATAC | 215365 | 1.5767692 | 5.398935 | 5 |
| TTGAG | 131780 | 1.5711527 | 5.531803 | 9 |
| GAGTC | 87145 | 1.5291787 | 8.141672 | 9 |
| AAGAC | 120750 | 1.4062037 | 5.510574 | 5 |
| CCATG | 82975 | 1.371765 | 5.5625997 | 9 |
| GTCTT | 123610 | 1.3636448 | 5.7933393 | 1 |
| TATGA | 174300 | 1.3544837 | 5.673972 | 4 |
| TGAGT | 111320 | 1.3272173 | 6.7052145 | 8 |
| GACTT | 117470 | 1.3195093 | 6.829185 | 7 |
| TCATA | 177885 | 1.3023638 | 5.413967 | 2 |
| GTGTT | 109935 | 1.2872624 | 5.8992863 | 1 |
| GATTG | 107130 | 1.2772619 | 6.0520344 | 7 |
| ATGAG | 104655 | 1.2704766 | 5.208778 | 7 |
| GTGTA | 103740 | 1.2368444 | 10.37156 | 1 |
| GTATG | 101965 | 1.2156819 | 5.3872933 | 3 |
| GTTCT | 107135 | 1.1818955 | 5.4135365 | 1 |
| TGGAC | 65730 | 1.1533986 | 7.1122656 | 5 |
| GTCCA | 67385 | 1.114027 | 7.5194855 | 1 |
| ACATG | 97375 | 1.1137066 | 5.2512074 | 8 |
| GGACT | 62915 | 1.1040024 | 7.418535 | 6 |
| TGTAT | 133430 | 1.0183383 | 5.095952 | 2 |
| GTATA | 100350 | 0.77981883 | 6.869353 | 1 |