![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-103_TAAGGCGA-AAGGAGTA_L005_R2_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1015664 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 39 |
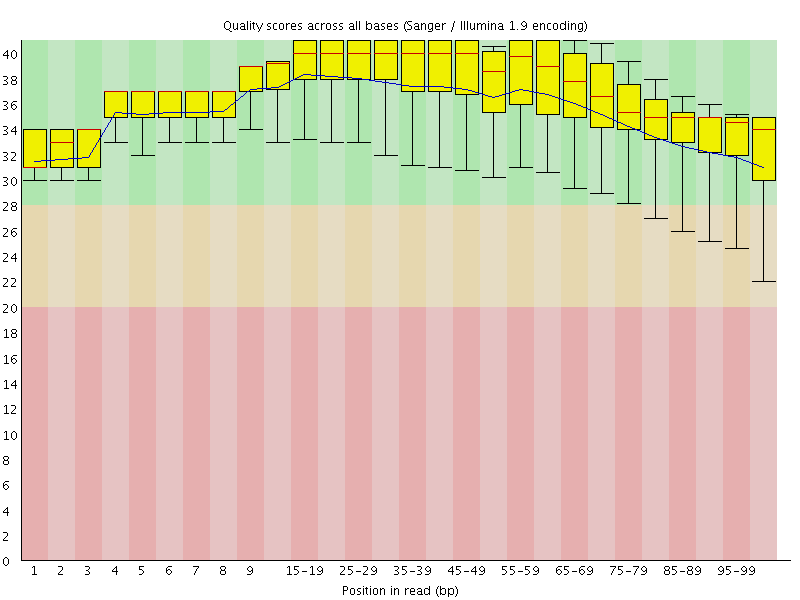
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
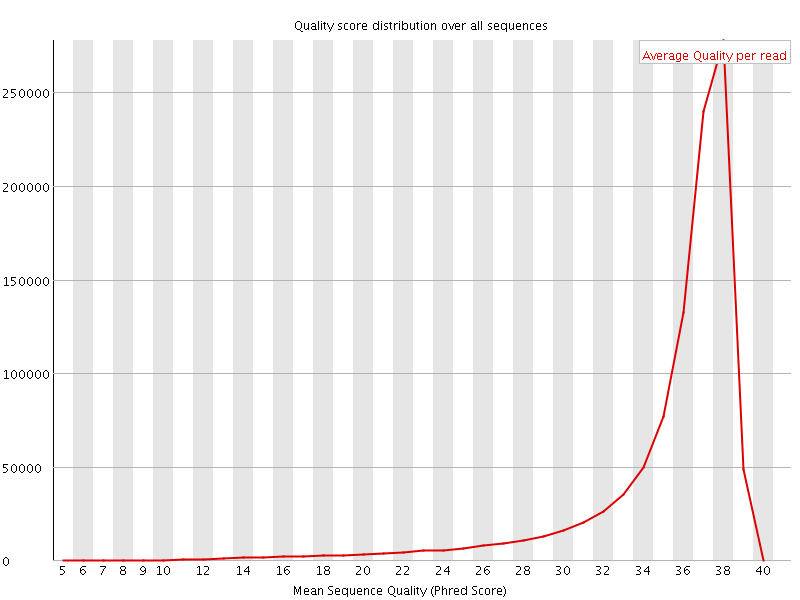
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
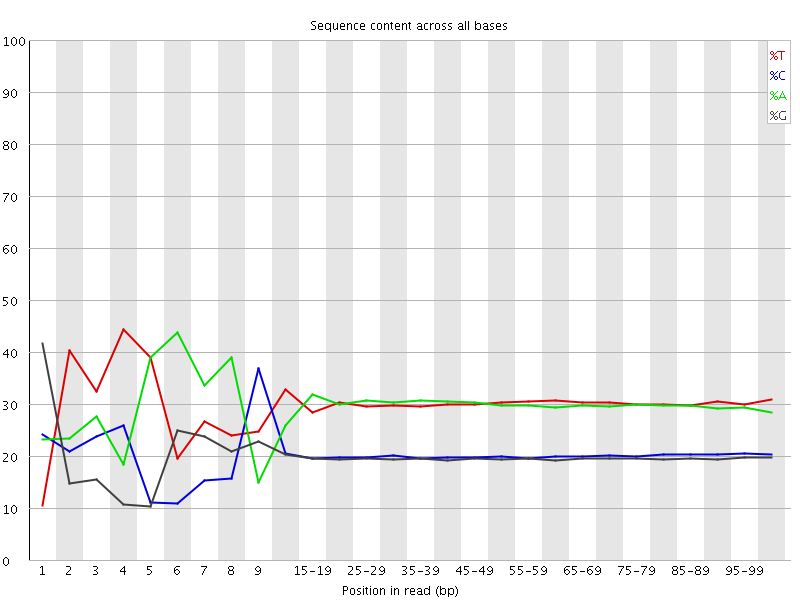
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
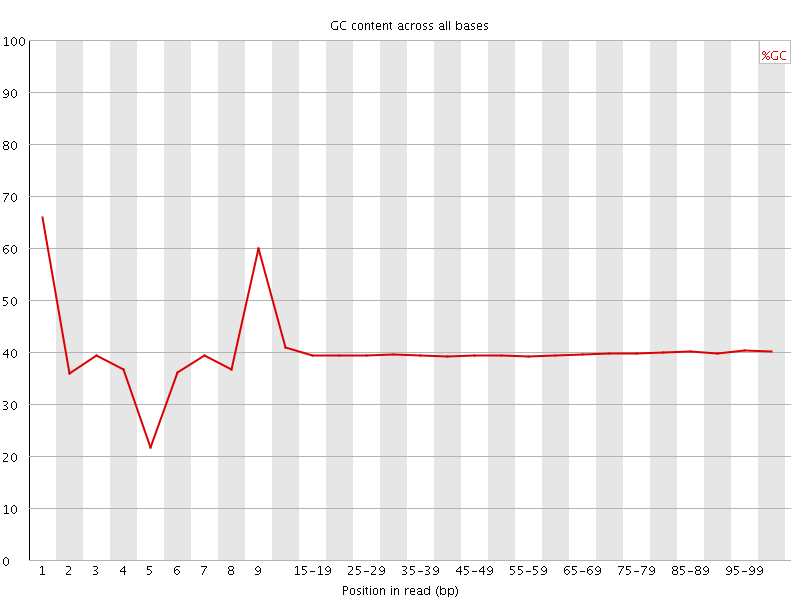
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
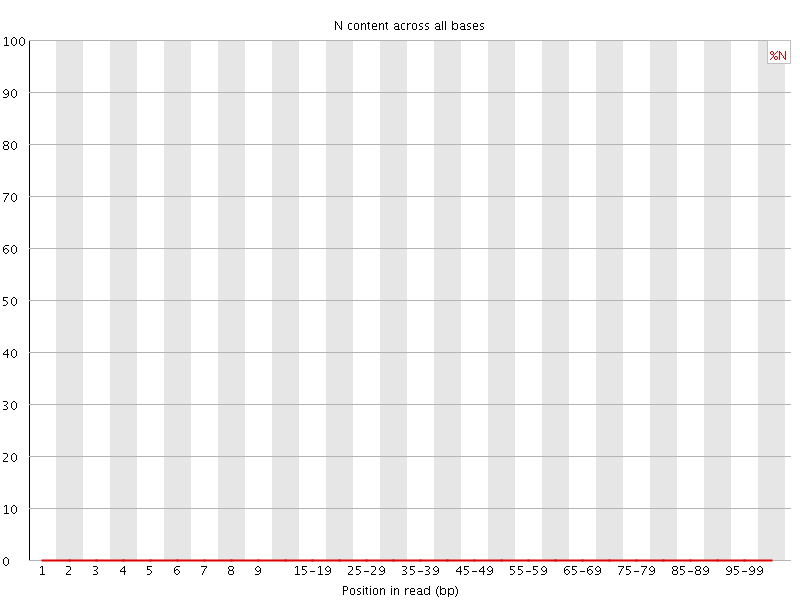
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
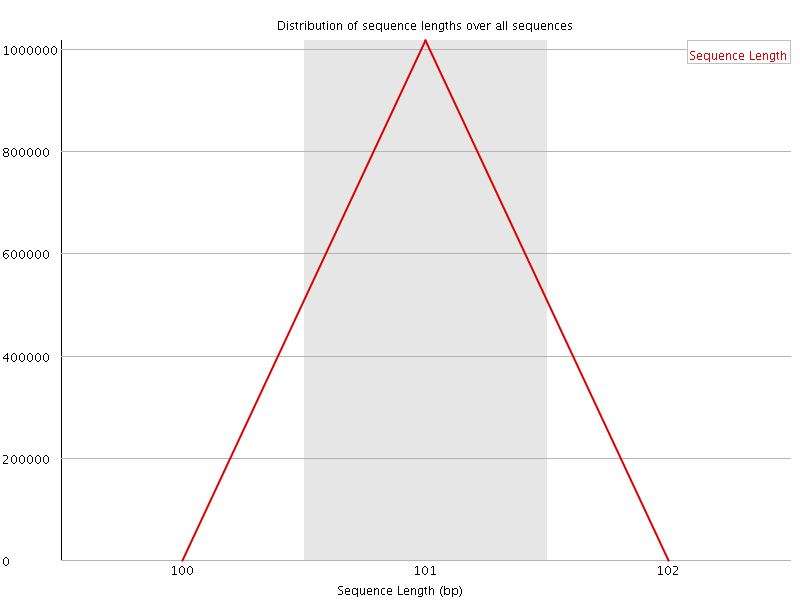
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
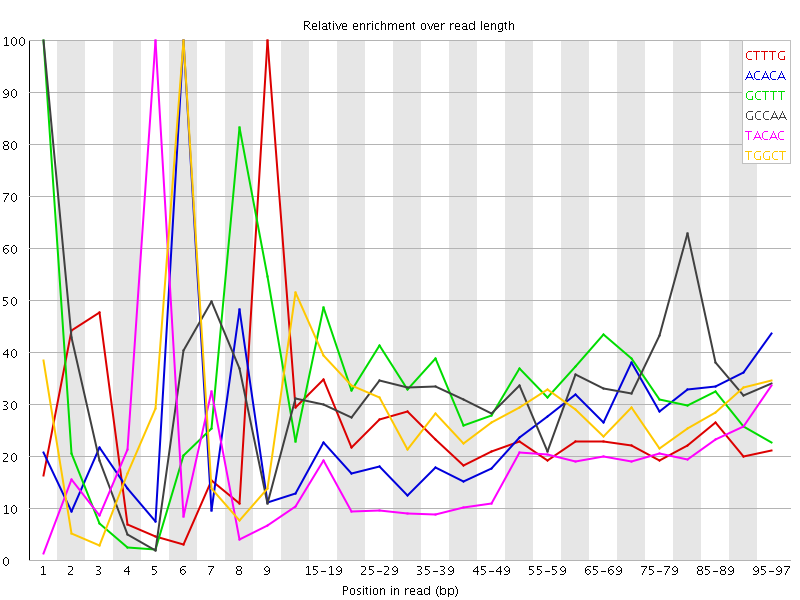
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CTTTG | 262245 | 2.4215124 | 10.057102 | 9 |
| ACACA | 226560 | 2.1252441 | 8.428917 | 6 |
| GCTTT | 220745 | 2.0383105 | 6.020157 | 1 |
| GCCAA | 133135 | 1.8845551 | 5.500196 | 1 |
| TACAC | 202840 | 1.8799819 | 10.808134 | 5 |
| TGGCT | 127340 | 1.7958119 | 6.0453167 | 6 |
| TTGGC | 126340 | 1.7817093 | 5.34273 | 5 |
| GACTC | 123860 | 1.7322961 | 6.443214 | 7 |
| CACAT | 184570 | 1.7106498 | 6.049688 | 7 |
| GAGTC | 117260 | 1.673676 | 9.752504 | 9 |
| CACCA | 120120 | 1.6661012 | 5.0446987 | 7 |
| TTGAG | 174020 | 1.6597193 | 6.2253556 | 9 |
| GGCTT | 117020 | 1.650274 | 5.0555625 | 1 |
| GAGAG | 111675 | 1.6463927 | 5.633672 | 7 |
| AAGAC | 161155 | 1.5427651 | 6.134442 | 5 |
| ATGGA | 158745 | 1.5323609 | 5.080506 | 4 |
| ATACA | 244305 | 1.5313408 | 5.8025146 | 6 |
| CATGA | 160310 | 1.516321 | 5.8665757 | 9 |
| CCATG | 106115 | 1.484116 | 5.914134 | 9 |
| GTGTA | 154265 | 1.4713054 | 12.117135 | 1 |
| GACTT | 156685 | 1.464308 | 7.940119 | 7 |
| GTCTT | 157700 | 1.456167 | 6.8040314 | 1 |
| TATGA | 229295 | 1.4492358 | 7.0976024 | 4 |
| TCCAT | 157815 | 1.4451829 | 5.734783 | 2 |
| ACTCA | 153705 | 1.4245839 | 5.726108 | 8 |
| TCATA | 229955 | 1.4241537 | 6.969766 | 2 |
| TGAGT | 148990 | 1.4209952 | 7.659243 | 8 |
| ATGAG | 138775 | 1.3395909 | 6.207152 | 7 |
| GTGTT | 142020 | 1.3383183 | 6.2535276 | 1 |
| AACAC | 140800 | 1.3207731 | 5.2829194 | 5 |
| GATTG | 136230 | 1.2992964 | 6.595427 | 7 |
| ACACC | 90575 | 1.2563031 | 5.616211 | 6 |
| GTTCT | 135995 | 1.2557477 | 5.442329 | 1 |
| TGGAC | 87780 | 1.2529019 | 8.363819 | 5 |
| GTCCA | 87195 | 1.2195024 | 9.63404 | 1 |
| ACATG | 125495 | 1.1870171 | 6.2565503 | 8 |
| TATAC | 191065 | 1.1833007 | 5.978377 | 5 |
| GGACT | 82375 | 1.1757553 | 8.720931 | 6 |
| ACACT | 125135 | 1.1597885 | 5.788775 | 6 |
| AGACT | 114835 | 1.0861874 | 5.9994164 | 6 |
| TGTAT | 171925 | 1.0736383 | 5.6707563 | 2 |
| CCATA | 115830 | 1.0735471 | 5.0533376 | 3 |
| GTATG | 112460 | 1.0725895 | 6.5232873 | 3 |
| GAGTA | 107325 | 1.0360051 | 5.235183 | 1 |
| CATAC | 110830 | 1.0272056 | 5.9614997 | 3 |
| GAATA | 144630 | 0.9251844 | 5.023956 | 1 |
| GTATA | 126965 | 0.8024694 | 7.551582 | 1 |