![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Nextera-103_TAAGGCGA-AAGGAGTA_L005_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 1015664 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
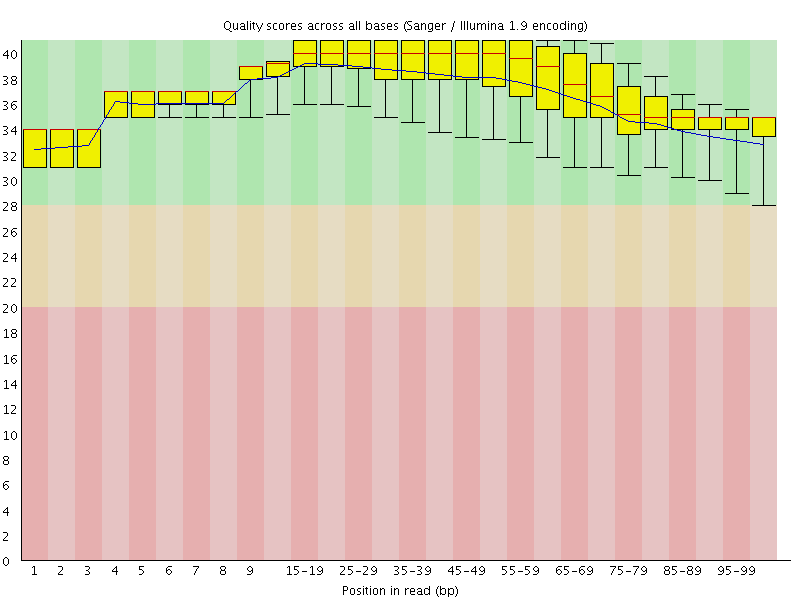
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
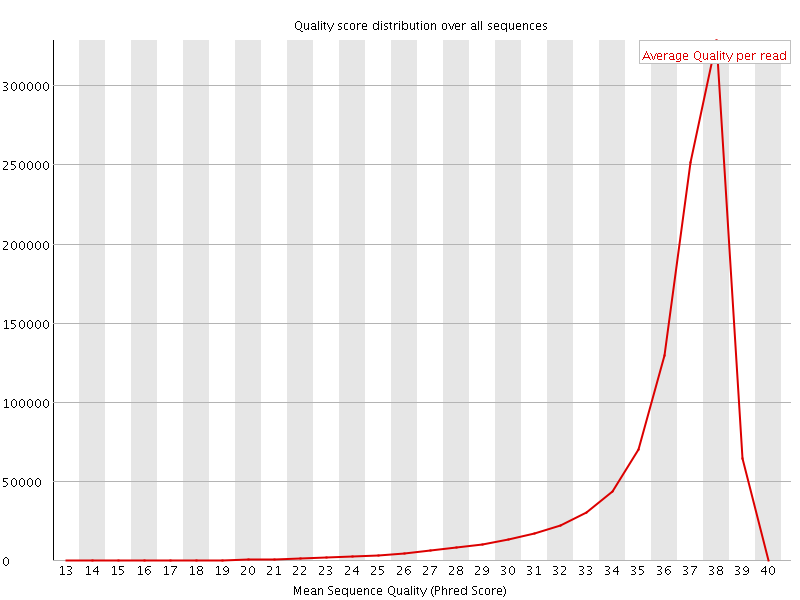
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
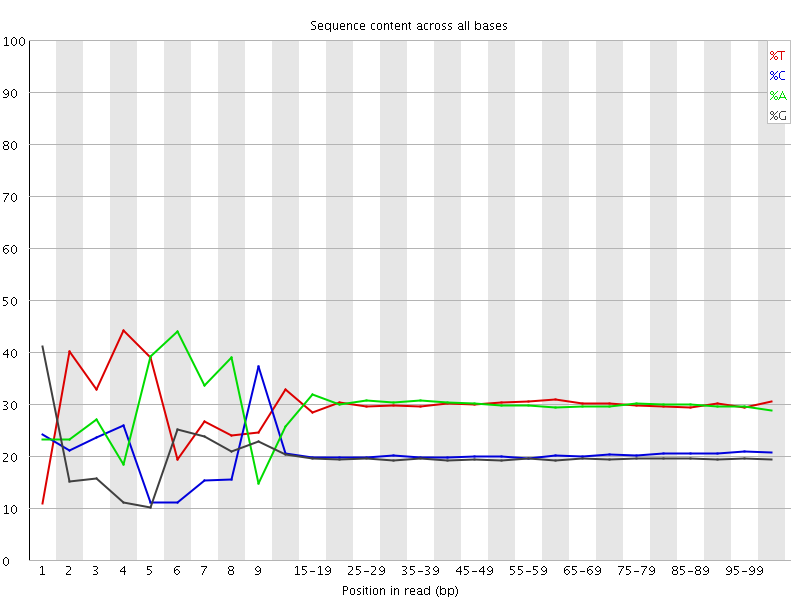
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
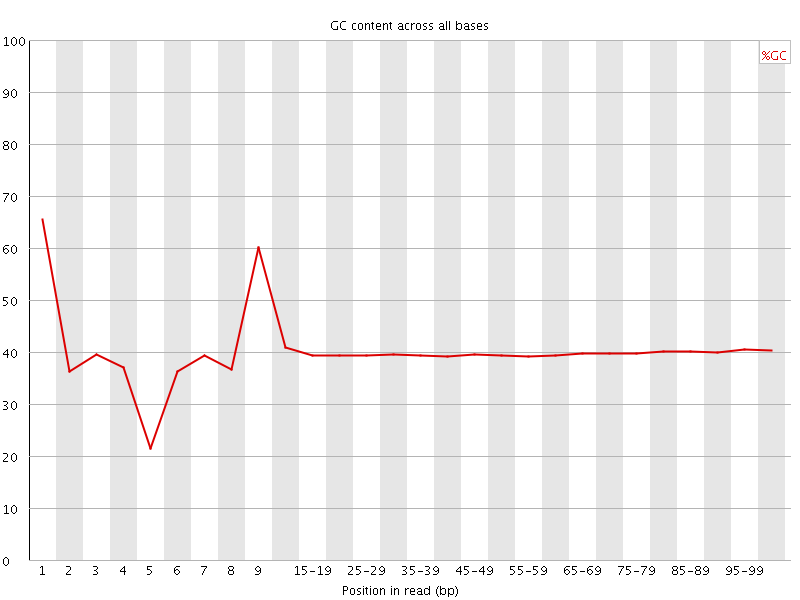
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content
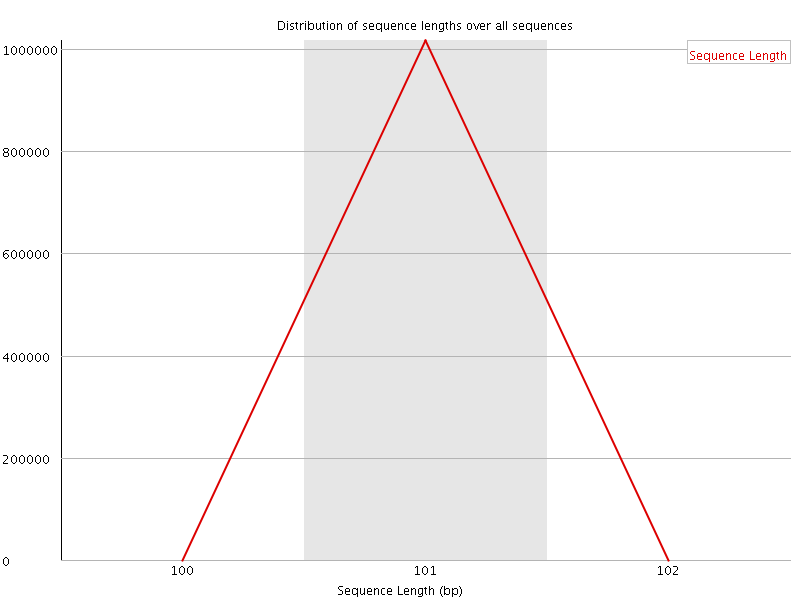
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
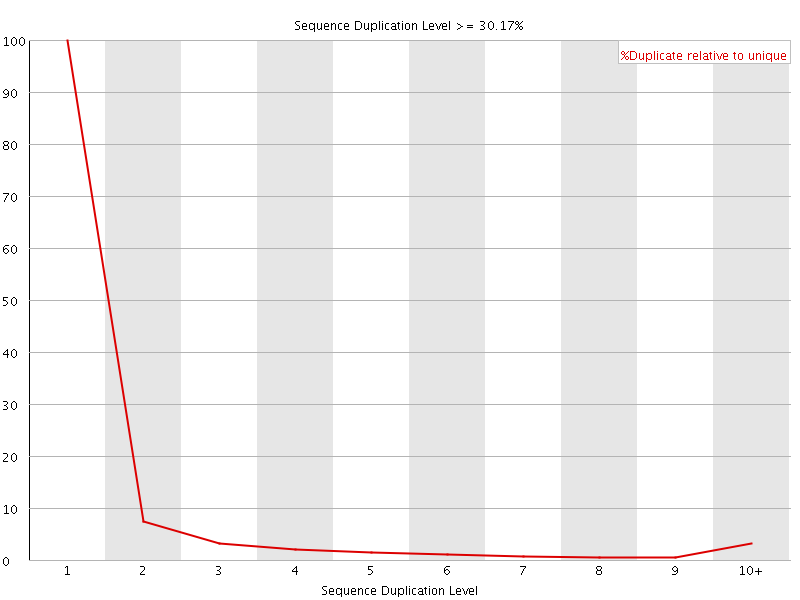
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CATATGGACTTTGGCTACACCATGAAAGCTTTGAGAAGCAAGAAGAAGGT | 1057 | 0.10406984987161108 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
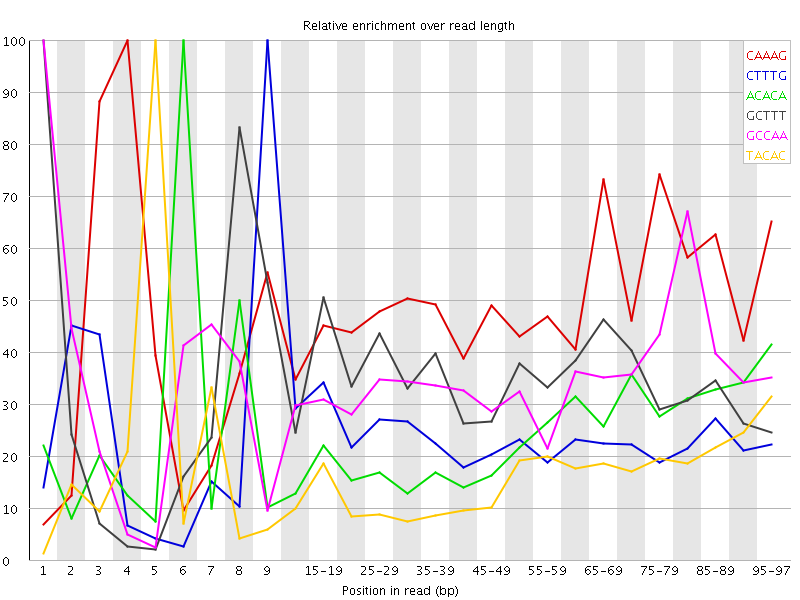
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CAAAG | 268165 | 2.5479894 | 5.1456475 | 4 |
| CTTTG | 264695 | 2.4637983 | 10.338937 | 9 |
| ACACA | 227900 | 2.0966682 | 8.657695 | 6 |
| GCTTT | 221225 | 2.0591767 | 5.922023 | 1 |
| GCCAA | 133965 | 1.8713688 | 5.3108125 | 1 |
| TACAC | 203900 | 1.8630472 | 11.3185005 | 5 |
| TGGCT | 127755 | 1.8180202 | 5.6368203 | 6 |
| TGGAG | 120355 | 1.7810403 | 5.0366077 | 5 |
| GACTC | 125485 | 1.7409294 | 6.4371347 | 7 |
| CACAT | 187880 | 1.7166717 | 6.3968363 | 7 |
| GAGTC | 118540 | 1.6984954 | 9.878486 | 9 |
| TTGAG | 171490 | 1.6599197 | 6.128904 | 9 |
| GAGAG | 111200 | 1.6568878 | 5.9237056 | 7 |
| GGCTT | 115990 | 1.6505982 | 5.555121 | 1 |
| AAGAC | 161255 | 1.5321761 | 6.1176543 | 5 |
| CATGA | 162045 | 1.5291582 | 6.258846 | 9 |
| ATACA | 245270 | 1.5243285 | 5.945009 | 6 |
| ATGGA | 155820 | 1.5186236 | 5.14572 | 4 |
| GTCTT | 161780 | 1.5058588 | 6.6171393 | 1 |
| CCATG | 107470 | 1.4909964 | 6.0201 | 9 |
| GACTT | 156010 | 1.4621454 | 8.024541 | 7 |
| TGAGT | 150820 | 1.4598464 | 7.6587825 | 8 |
| TATGA | 228265 | 1.4551404 | 6.938622 | 4 |
| TCATA | 235415 | 1.4530802 | 7.207569 | 2 |
| TCCAT | 159370 | 1.4462206 | 5.522664 | 2 |
| TGTGT | 149915 | 1.4411681 | 5.0616336 | 8 |
| ACTCA | 154800 | 1.4144174 | 5.7146244 | 8 |
| AGACT | 148555 | 1.4018582 | 6.2222447 | 6 |
| GTGTT | 141570 | 1.3609457 | 6.1348276 | 1 |
| ATGAG | 139255 | 1.3571808 | 6.350641 | 7 |
| GTGTA | 138450 | 1.3401124 | 12.156959 | 1 |
| GATTG | 136185 | 1.3181884 | 6.692049 | 7 |
| AACAC | 141245 | 1.2994466 | 5.263307 | 5 |
| GTTCT | 135380 | 1.2601259 | 5.50676 | 1 |
| GTCCA | 89240 | 1.2380806 | 9.526416 | 1 |
| TGGAC | 86295 | 1.2364744 | 8.766983 | 5 |
| ACACC | 90975 | 1.2304953 | 5.6986117 | 6 |
| ACCAT | 132045 | 1.2065037 | 5.1032934 | 8 |
| ACATG | 127445 | 1.202651 | 6.601985 | 8 |
| GTATG | 122800 | 1.1886297 | 6.6732774 | 3 |
| TATAC | 192335 | 1.1871724 | 6.2574997 | 5 |
| ATGTG | 120375 | 1.1651572 | 5.0964694 | 7 |
| GGACT | 80975 | 1.1602468 | 8.753089 | 6 |
| ACACT | 124990 | 1.1420417 | 5.891823 | 6 |
| TGTAT | 172115 | 1.0896965 | 5.8978148 | 2 |
| CCATA | 116060 | 1.0604477 | 5.0634236 | 3 |
| GAGTA | 108405 | 1.0565164 | 5.075842 | 1 |
| CATAC | 113140 | 1.0337673 | 6.277227 | 3 |
| GGATA | 92365 | 0.90019035 | 5.2837906 | 1 |
| ATACT | 132595 | 0.8184319 | 5.4943895 | 6 |
| GTATA | 126545 | 0.8066972 | 7.140924 | 1 |