![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 3L15C_GCCAAT_L004_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 18359396 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 46 |
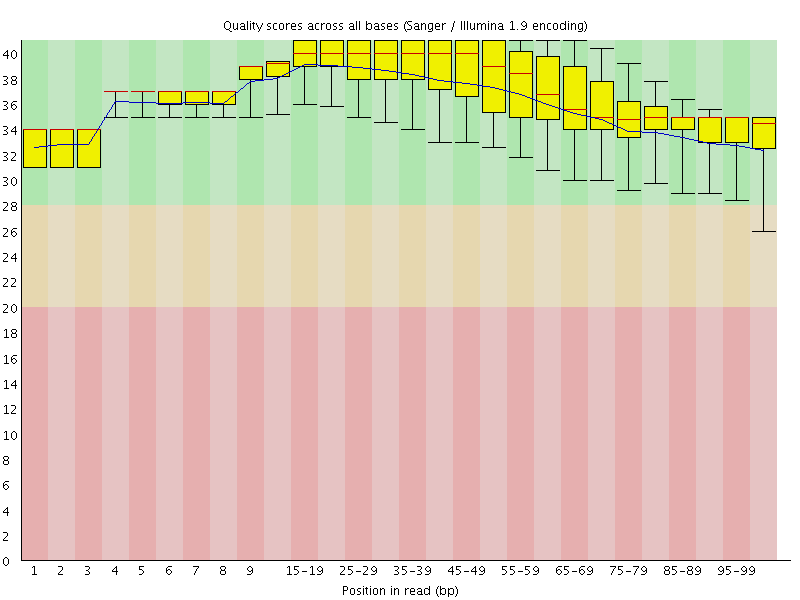
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
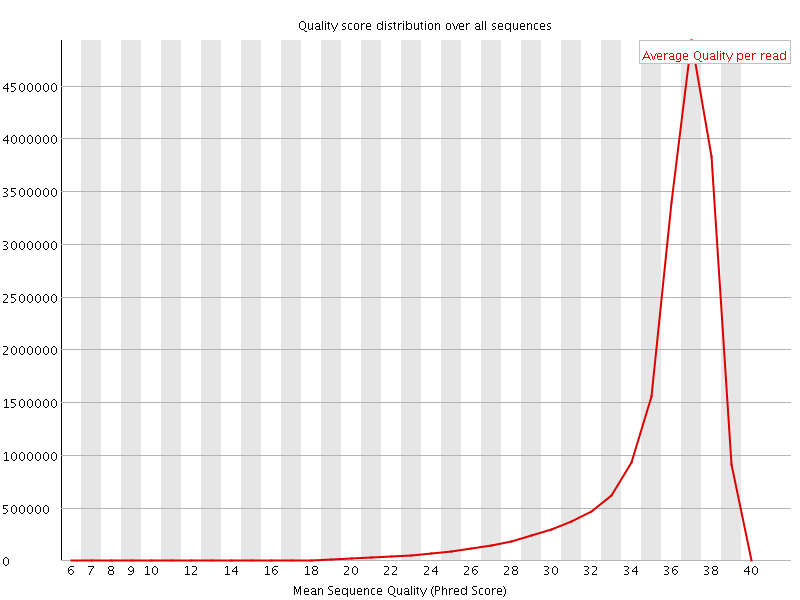
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
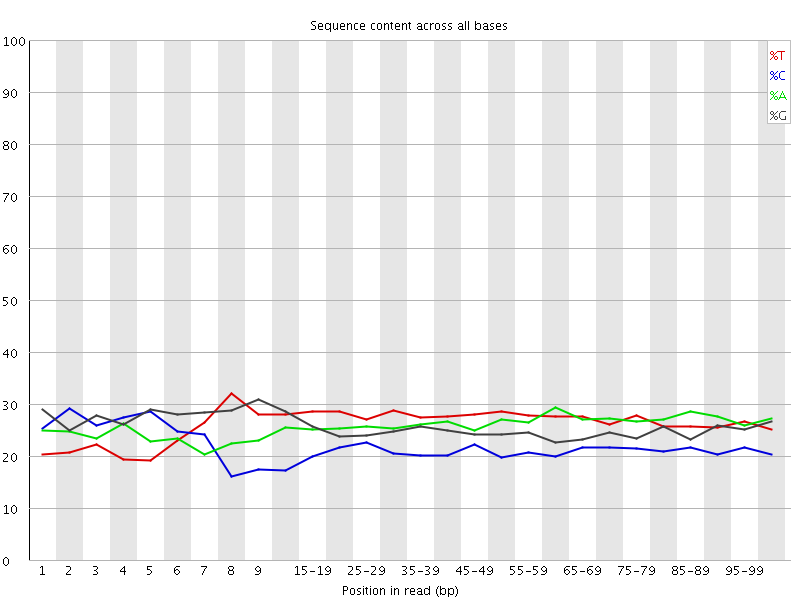
![[OK]](Icons/tick.png) Per base sequence content
Per base sequence content
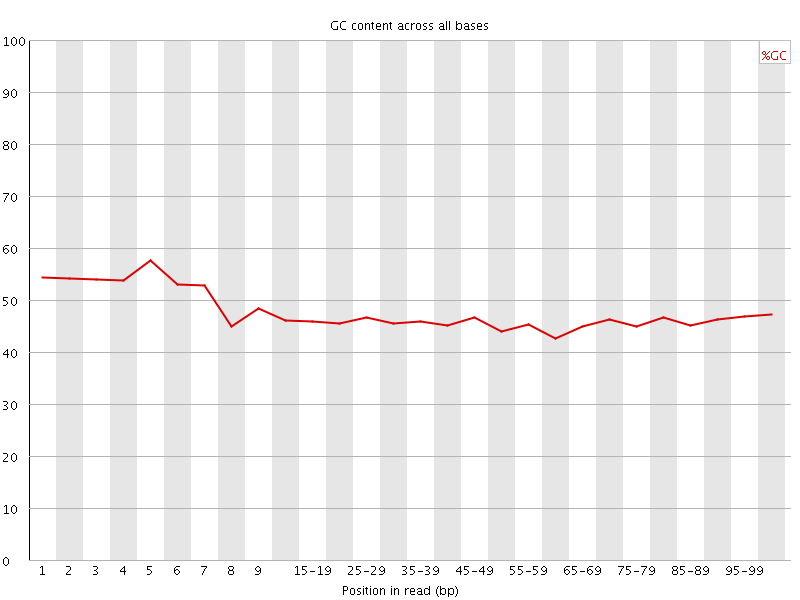
![[WARN]](Icons/warning.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
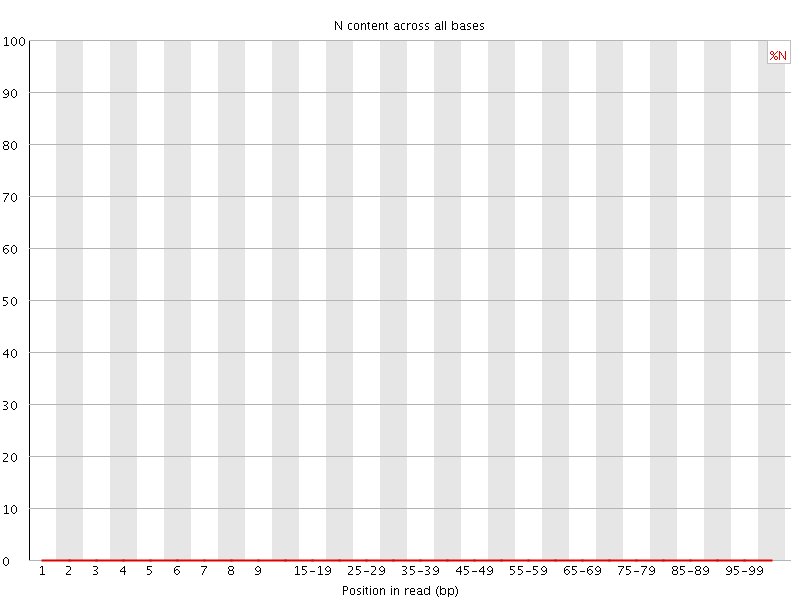
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
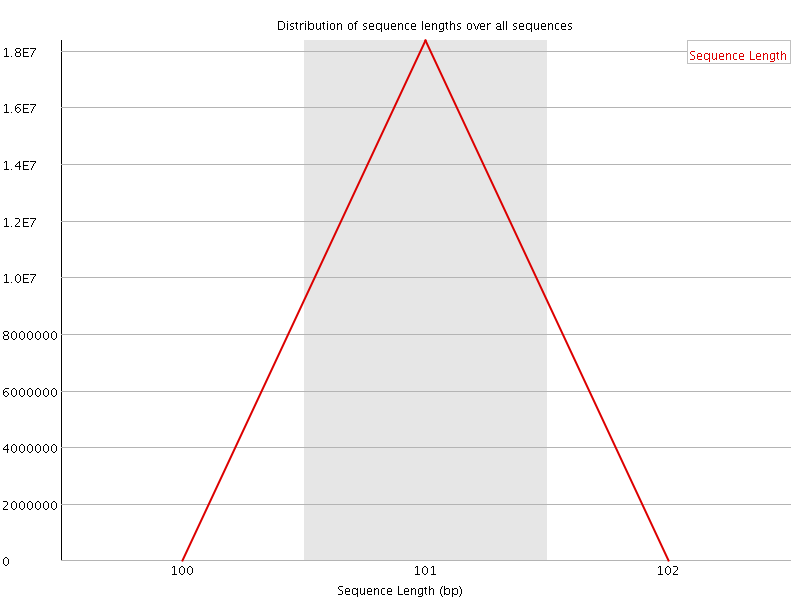
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GCGAGTCGGGCTGTTTGGGAATGCAGCCCTAAGTGGGAGGTAAATTCCTT | 376940 | 2.0531176515828737 | No Hit |
| GATACCGTCCTAGTCTCAACCATAAACGATGCCGACTAGGGATTGGCGGA | 261603 | 1.4248998169656562 | No Hit |
| ACCCAATTGGTCAAGGAAGCTTCTCTGACGGTATGCCTTTAGGAATCTCT | 218126 | 1.188089194219679 | No Hit |
| CTGGTACTTTCAACTTCATGATTGTGTTCCAAGCTGAACACAACATCCTT | 144313 | 0.7860443774947715 | No Hit |
| CAGACCTCTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAA | 112932 | 0.6151182751327985 | No Hit |
| GGTACTTTCAACTTCATGATTGTGTTCCAAGCTGAACACAACATCCTTAT | 87058 | 0.47418771292911815 | No Hit |
| GGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAAC | 84490 | 0.4602003246729903 | No Hit |
| CCCCGGTTGTTACCCGACTACCAATAGGGGTGCTTCGTGCACCCCCAGAA | 84004 | 0.45755317876470447 | No Hit |
| AGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAG | 59944 | 0.3265031158977126 | No Hit |
| GACTGCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTCT | 59471 | 0.32392677841907214 | No Hit |
| TGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTT | 58604 | 0.3192044008419449 | No Hit |
| GTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAACTTC | 58164 | 0.31680780783855855 | No Hit |
| CTGACGGTATGCCTTTAGGAATCTCTGGTACTTTCAACTTCATGATTGTG | 57677 | 0.3141552151279922 | No Hit |
| CAACACGGGGAAACTTACCAGGTCCAGACATAGTAAGGATTGACAGATTG | 56559 | 0.3080656901784786 | No Hit |
| CATGGAGAGTTTGATCCTGGCTCAGGATGAACGCTGGCGGCATGCTTAAC | 56402 | 0.30721054222045213 | No Hit |
| GATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAAGCGGAGGAG | 55618 | 0.30294024923260005 | No Hit |
| GCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTCTGCGA | 53099 | 0.289219754288213 | No Hit |
| CAAACGAATAAGGGCTTATGGTGGATACCTAGGCACCCAGAGACGAGGAA | 52862 | 0.2879288621477526 | No Hit |
| TGCCAGTAGTCATATGCTTGTCTCAAAGATTAAGCCATGCATGTGTAAGT | 52470 | 0.2857937156538265 | No Hit |
| ACATGGGTCGTGAGTGGGAACTTAGCTACCGTTTAGGTATGCGTCCTTGG | 50430 | 0.27468223900176236 | No Hit |
| ACCTCCCCGGTTGTTACCCGACTACCAATAGGGGTGCTTCGTGCACCCCC | 48648 | 0.26497603733804753 | No Hit |
| GCTACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGGAAGCTTCTCTGA | 48626 | 0.26485620768787815 | No Hit |
| CTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAAGCGGAGG | 48123 | 0.2621164661408251 | No Hit |
| GGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTC | 40896 | 0.2227524260602037 | No Hit |
| GTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTT | 39041 | 0.21264860783001793 | No Hit |
| TGGGTCGGGGGGAGCGGTCCGCCCCTTGTGGGTGTGCACTGGTCCACTCG | 36559 | 0.19912964457000654 | No Hit |
| GCAGACCTCTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTA | 35858 | 0.1953114361714296 | No Hit |
| ATGCATGGTTCCTTAGTAACTTCAAGTTTAATCCGTGAAACTACTGAGAA | 35247 | 0.19198343997809078 | No Hit |
| CCTGTAGATATTGATGGTATCCGTGAGCCTGTTTCTGGTTCTCTTCTTTA | 34268 | 0.18665102054555607 | No Hit |
| GAACACAACATCCTTATGCACCCATTCCACATGCTTGGTGTAGCTGGTGT | 30918 | 0.16840423290613699 | No Hit |
| GCAACTTCTGTATTTATTATTGCTTTCATTGCAGCTCCTCCTGTAGATAT | 28903 | 0.15742892631108343 | No Hit |
| GCTTGGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGCACTAT | 28656 | 0.1560835661478188 | No Hit |
| GAAACATCTTAGTAGCCAGAGGAAAAGAAAGCAAAAGCGATTCCCATAGT | 28532 | 0.15540816266504628 | No Hit |
| TGACTGCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTC | 28446 | 0.15493973766892985 | No Hit |
| GCTTGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATG | 26119 | 0.14226502876238414 | No Hit |
| TGCTACATGGGTCGTGAGTGGGAACTTAGCTACCGTTTAGGTATGCGTCC | 26074 | 0.14201992265976507 | No Hit |
| TGGCAAGAGACCGATAGCGAACAAGTACCGCGAGGGAAAGATGAAAAGGA | 25089 | 0.13665482241354782 | No Hit |
| AAGGCTAAATATTGGCAAGAGACCGATAGCGAACAAGTACCGCGAGGGAA | 25052 | 0.13645329072917214 | No Hit |
| CCACGAGGCGCGATCTGCGAGTCGGGCTGTTTGGGAATGCAGCCCTAAGT | 24832 | 0.13525499422747894 | No Hit |
| ACCTGGTTGATCCTGCCAGTAGTCATATGCTTGTCTCAAAGATTAAGCCA | 24444 | 0.13314163494267459 | No Hit |
| AGCAAGCGACGAAATGCTTCGGGGAGCTGAAAATAAGCATAGATCCGGAG | 24402 | 0.1329128692468968 | No Hit |
| AACACGGACCAAGGAGTCTAACATGTATGCAAGCAGGTGGGCGGCAAACC | 24153 | 0.13155661547907133 | No Hit |
| TGGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGCACTATGGC | 23506 | 0.12803253440363724 | No Hit |
| AGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCGCCT | 23490 | 0.12794538556715046 | No Hit |
| CTAAGGCTAAATATTGGCAAGAGACCGATAGCGAACAAGTACCGCGAGGG | 22866 | 0.12454658094416614 | No Hit |
| AAGTGGGAGGTAAATTCCTTCTAAGGCTAAATATTGGCAAGAGACCGATA | 22707 | 0.12368053938157877 | No Hit |
| CCACATGCTTGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTA | 22326 | 0.12160530771273739 | No Hit |
| CAACGCGAAGAACCTTACCAGGGCTTGACATGCCGCGAATCCTTTTGAAA | 21999 | 0.11982420336703888 | No Hit |
| TAGTAGCCAGAGGAAAAGAAAGCAAAAGCGATTCCCATAGTAGCGGCGAG | 21855 | 0.11903986383865788 | No Hit |
| ACCAATAGGGGTGCTTCGTGCACCCCCAGAAGCATGTGTCGAGAGCTCCC | 21211 | 0.11553212317006509 | No Hit |
| ACAGACCTCTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTA | 21012 | 0.11444820951626077 | No Hit |
| AAATCCCTTAACGAGGATCCATTGGAGGGCAAGTCTGGTGCCAGCAGCCG | 20823 | 0.11341876388526072 | No Hit |
| CTATTCAGTGCTATGCATGGTTCCTTAGTAACTTCAAGTTTAATCCGTGA | 20676 | 0.11261808395003844 | No Hit |
| CCAGACCTCTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTA | 20486 | 0.11158319151675795 | No Hit |
| GCTCCACGAGGCGCGATCTGCGAGTCGGGCTGTTTGGGAATGCAGCCCTA | 20314 | 0.1106463415245251 | No Hit |
| CTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTA | 20308 | 0.11061366071084255 | No Hit |
| CCTGTTGCGGCTGCTACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGG | 20080 | 0.10937178979090598 | No Hit |
| ACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGGAAGCTTCTCTGACGG | 20044 | 0.10917570490881073 | No Hit |
| ACATGCTTGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATG | 20024 | 0.10906676886320225 | No Hit |
| GTAGGTGGCTTTTTAAGTCTGCTGTCAAATCCCAGGGCTCAACCCTGGAC | 19634 | 0.10694251597383704 | No Hit |
| GTATTTATTATTGCTTTCATTGCAGCTCCTCCTGTAGATATTGATGGTAT | 19523 | 0.10633792092071004 | No Hit |
| TGGTGGTAGTAGCAAATATTCAAATGAGAACTTTGAAGACTGAAGTGGAG | 18987 | 0.10341843489840297 | No Hit |
| GCAGCGTGCCGGCCCCAAGTTCCCTGGAATGGGATGCCCAAGAGGGTGAC | 18699 | 0.10184975584164098 | No Hit |
| CAAGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCGC | 18640 | 0.101528394507096 | No Hit |
| GGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAACTTCAAGTTTAAT | 18623 | 0.10143579886832878 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
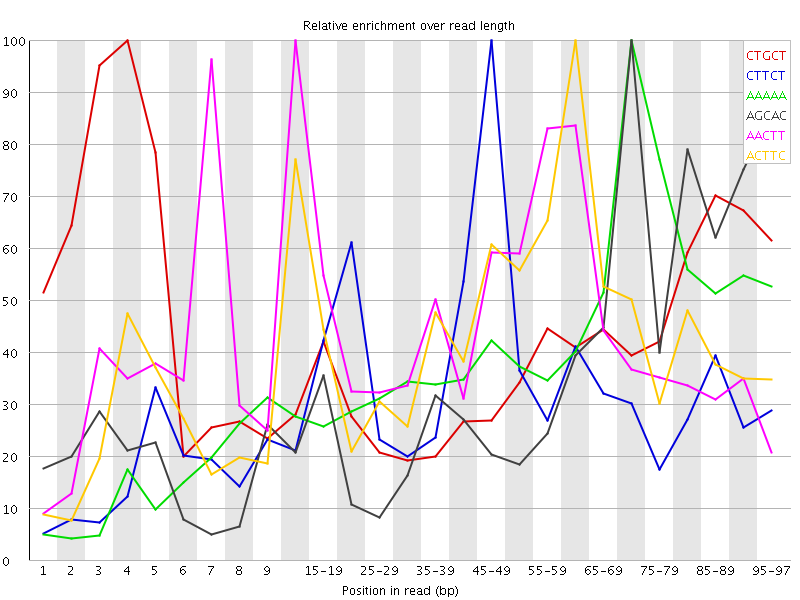
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CTGCT | 4667730 | 3.1304262 | 7.691796 | 4 |
| CTTCT | 4889720 | 3.0381634 | 8.801376 | 45-49 |
| AAAAA | 6723725 | 2.9281385 | 6.9136353 | 70-74 |
| AGCAC | 3841520 | 2.7208922 | 7.144012 | 70-74 |
| AACTT | 5212960 | 2.6749222 | 5.6773295 | 10-14 |
| ACTTC | 4187250 | 2.6736913 | 5.8710203 | 60-64 |
| TCCTT | 4036205 | 2.5078428 | 10.285098 | 45-49 |
| TCTCT | 4030720 | 2.504435 | 7.753425 | 20-24 |
| TGAAA | 5567150 | 2.4778478 | 5.671092 | 65-69 |
| GAAGA | 5133660 | 2.4662633 | 5.1923976 | 90-94 |
| CTGGT | 4331825 | 2.4520447 | 12.070883 | 1 |
| CGGAA | 4046815 | 2.419253 | 5.974427 | 95-97 |
| AAGAG | 4969050 | 2.387183 | 5.4122515 | 65-69 |
| ATCGG | 4062500 | 2.3632302 | 7.350353 | 95-97 |
| CTGAA | 4265360 | 2.362398 | 5.8252196 | 80-84 |
| TGGTG | 4788545 | 2.2878144 | 7.3810687 | 4 |
| AGCTG | 3927470 | 2.2846808 | 5.525209 | 1 |
| AGAGC | 3766660 | 2.2517717 | 7.0298634 | 95-97 |
| GAAAC | 3906915 | 2.2237532 | 5.0386434 | 60-64 |
| GGTGG | 4305245 | 2.2201672 | 6.0977736 | 7 |
| TCGGA | 3786660 | 2.2027693 | 6.2616596 | 95-97 |
| CTACT | 3447045 | 2.201047 | 8.974966 | 2 |
| AGATC | 3937275 | 2.1806858 | 8.172 | 95-97 |
| TGCTA | 3989990 | 2.1503735 | 5.3966208 | 4 |
| ATGCA | 3851470 | 2.1331623 | 5.6421857 | 20-24 |
| TTCAA | 4151860 | 2.1304407 | 8.700282 | 9 |
| CTTCA | 3323410 | 2.1221023 | 6.045298 | 60-64 |
| GCAGC | 2818550 | 2.0967627 | 7.868787 | 20-24 |
| TCAAC | 3173880 | 2.082707 | 6.766465 | 8 |
| GATCG | 3568810 | 2.076042 | 7.7427115 | 95-97 |
| CTCTG | 3063980 | 2.0548668 | 8.561178 | 6 |
| TTCCT | 3294520 | 2.0470068 | 8.651045 | 45-49 |
| ATTCC | 3195065 | 2.04015 | 6.2167444 | 40-44 |
| TATGC | 3757645 | 2.025153 | 5.83193 | 30-34 |
| CCTTA | 3117690 | 1.9907435 | 6.369351 | 95-97 |
| CGGTG | 3231365 | 1.9743042 | 6.7260866 | 9 |
| GGTGT | 4063880 | 1.9415926 | 6.8376284 | 1 |
| GTCTG | 3415590 | 1.9334065 | 5.7509756 | 90-94 |
| CAAGC | 2728395 | 1.932482 | 5.330308 | 30-34 |
| GAAAG | 3931135 | 1.8885579 | 5.273203 | 15-19 |
| TTTCA | 3778745 | 1.8867705 | 9.497133 | 8 |
| CACAC | 2247550 | 1.8860701 | 7.345564 | 95-97 |
| CTGTT | 3561140 | 1.8675659 | 5.89291 | 10-14 |
| GCACC | 2114535 | 1.8637117 | 6.8866606 | 70-74 |
| CATGG | 3199395 | 1.8611465 | 6.381966 | 2 |
| CAACT | 2823530 | 1.8528064 | 7.7621446 | 9 |
| AGGAA | 3834405 | 1.8420877 | 5.6218047 | 10-14 |
| CAAGT | 3303045 | 1.8294134 | 5.0179434 | 85-89 |
| GCTGC | 2523670 | 1.8268416 | 5.508312 | 3 |
| GAGCA | 3032145 | 1.8126663 | 5.310129 | 95-97 |
| GCAAG | 3015915 | 1.802964 | 5.743796 | 65-69 |
| CCCTA | 2189855 | 1.7881691 | 7.4719152 | 25-29 |
| CAGCA | 2523635 | 1.7874535 | 5.0624948 | 70-74 |
| GCTTC | 2653060 | 1.7792821 | 6.2273784 | 15-19 |
| GCACA | 2504010 | 1.7735535 | 6.440769 | 95-97 |
| ACTTT | 3550020 | 1.7725656 | 9.1836605 | 6 |
| GAGAA | 3666705 | 1.761523 | 5.2060804 | 80-84 |
| CTTTC | 2822660 | 1.753823 | 10.658165 | 7 |
| CTAAG | 3163605 | 1.7521836 | 5.565021 | 50-54 |
| GCGAA | 2915155 | 1.742728 | 5.58689 | 80-84 |
| GCTGG | 2828900 | 1.7284056 | 6.734999 | 2 |
| ACCCA | 2058610 | 1.7275181 | 22.348833 | 1 |
| CAGCC | 1959065 | 1.7266834 | 7.870916 | 20-24 |
| TGGGA | 3501265 | 1.7190852 | 6.0314713 | 30-34 |
| TGCAG | 2950705 | 1.716479 | 6.3402042 | 20-24 |
| CTACC | 2091745 | 1.7080555 | 7.7898145 | 20-24 |
| CCAGC | 1935030 | 1.7054994 | 5.796368 | 70-74 |
| GGAGG | 3217520 | 1.7051567 | 6.000957 | 35-39 |
| CCGCC | 1531365 | 1.6795703 | 7.1069303 | 65-69 |
| TACTT | 3348645 | 1.6720169 | 9.044199 | 5 |
| CACCC | 1597760 | 1.6684594 | 5.8716545 | 50-54 |
| ACTCC | 2028360 | 1.6562973 | 5.394018 | 60-64 |
| ATGCC | 2401220 | 1.6549503 | 5.3787327 | 30-34 |
| GCGAG | 2629470 | 1.6510168 | 25.578794 | 1 |
| TTGGT | 3713930 | 1.6439188 | 13.790634 | 7 |
| GGAAA | 3412395 | 1.6393499 | 5.195985 | 9 |
| CAACA | 2425640 | 1.6357588 | 5.5293083 | 1 |
| TCTGA | 3034860 | 1.6356138 | 6.9475102 | 7 |
| TGTTT | 3978355 | 1.6314716 | 5.307481 | 10-14 |
| CCAAT | 2426555 | 1.5923104 | 17.104532 | 3 |
| TTCGG | 2795675 | 1.5825015 | 5.863507 | 7 |
| ACACG | 2218135 | 1.5710723 | 5.7102714 | 95-97 |
| GAACA | 2730295 | 1.55404 | 8.59888 | 80-84 |
| GCTAA | 2784745 | 1.5423495 | 6.20331 | 55-59 |
| ACTGC | 2229055 | 1.536292 | 6.3430476 | 2 |
| GGGAA | 3037890 | 1.5328504 | 5.836843 | 15-19 |
| CAAGA | 2681680 | 1.5263691 | 5.331802 | 65-69 |
| GGAAT | 3233570 | 1.5116088 | 5.6711063 | 15-19 |
| CTCTA | 2363075 | 1.508898 | 5.472698 | 95-97 |
| TTTGG | 3357765 | 1.4862671 | 5.102679 | 95-97 |
| ACCGC | 1684765 | 1.4849205 | 6.8043275 | 90-94 |
| AGAGA | 3082205 | 1.4807231 | 5.2499127 | 70-74 |
| GCTGT | 2541415 | 1.4385768 | 5.965598 | 10-14 |
| GGGAG | 2709450 | 1.4358999 | 5.3308077 | 35-39 |
| AGCCC | 1622355 | 1.4299134 | 8.522568 | 25-29 |
| GCTAC | 2067140 | 1.4246982 | 8.979301 | 1 |
| ATTGG | 3052010 | 1.3883141 | 12.111101 | 6 |
| TTGGG | 2897525 | 1.3843453 | 5.015085 | 95-97 |
| GTGTA | 3036280 | 1.381159 | 6.3581953 | 2 |
| AGCGA | 2294850 | 1.3718992 | 5.5734973 | 80-84 |
| CAACC | 1625895 | 1.3643978 | 5.8129134 | 15-19 |
| TGGTA | 2999220 | 1.3643008 | 8.752442 | 2 |
| TCGGT | 2404790 | 1.3612398 | 6.29323 | 8 |
| CATCC | 1663980 | 1.3587556 | 5.161442 | 90-94 |
| CGCGA | 1825090 | 1.3577126 | 5.503605 | 90-94 |
| TGCCG | 1862915 | 1.3485323 | 5.626836 | 30-34 |
| GCCCT | 1561625 | 1.3393232 | 8.150632 | 25-29 |
| CGAGG | 2132210 | 1.3387924 | 6.445263 | 95-97 |
| CAGGA | 2225360 | 1.330357 | 5.3645263 | 4 |
| GTTAC | 2452745 | 1.3218875 | 5.9536695 | 9 |
| TACCC | 1616645 | 1.3201033 | 5.4992003 | 20-24 |
| CCCAA | 1570120 | 1.3175932 | 22.474136 | 2 |
| GAGTC | 2260015 | 1.3146919 | 23.227343 | 3 |
| ATCTC | 2024060 | 1.2924262 | 5.571855 | 45-49 |
| GGCTG | 2100350 | 1.283275 | 26.260735 | 9 |
| AGGGA | 2537650 | 1.2804404 | 5.203851 | 95-97 |
| TGGTC | 2224255 | 1.2590473 | 15.141455 | 8 |
| AACAC | 1858885 | 1.2535607 | 7.757312 | 2 |
| CCAGT | 1788110 | 1.2323873 | 6.035323 | 3 |
| ACGGT | 2117985 | 1.2320704 | 5.270546 | 4 |
| CCTTC | 1531340 | 1.2167732 | 7.1325774 | 45-49 |
| CCTCC | 1190885 | 1.2100929 | 6.4561896 | 2 |
| CCACA | 1440555 | 1.2088666 | 5.07951 | 60-64 |
| GGCAA | 2021790 | 1.2086595 | 5.3268423 | 65-69 |
| CGCCA | 1369000 | 1.206611 | 5.3364844 | 65-69 |
| GGGCT | 1943410 | 1.1873876 | 25.035704 | 8 |
| ACAAG | 2085595 | 1.187087 | 5.342925 | 85-89 |
| AGACC | 1667260 | 1.1808957 | 9.239467 | 2 |
| TACCG | 1713025 | 1.1806378 | 19.938215 | 3 |
| ACATG | 2110385 | 1.1688508 | 5.271317 | 1 |
| CGAAC | 1631775 | 1.1557622 | 5.7860956 | 80-84 |
| GCCAG | 1550310 | 1.1532996 | 6.405069 | 2 |
| AAGGC | 1928165 | 1.152689 | 5.503947 | 50-54 |
| AGGTA | 2452685 | 1.1465654 | 5.5081906 | 35-39 |
| CCTAG | 1662330 | 1.1456982 | 18.83478 | 9 |
| CCGAC | 1297360 | 1.1434689 | 5.38203 | 30-34 |
| CCGCG | 1222305 | 1.131511 | 6.8254523 | 90-94 |
| GAGGG | 2107835 | 1.1170679 | 5.4321375 | 95-97 |
| AATGC | 1996200 | 1.1056086 | 5.2296205 | 20-24 |
| AAGAC | 1927090 | 1.0968686 | 5.3959646 | 9 |
| CACCT | 1336690 | 1.0915005 | 6.0215807 | 70-74 |
| GACCG | 1431510 | 1.0649225 | 5.681161 | 70-74 |
| TCGGG | 1740775 | 1.0635813 | 24.92044 | 6 |
| CTAAA | 1991185 | 1.0500106 | 5.0098753 | 55-59 |
| CCCCG | 952070 | 1.0442111 | 11.416925 | 1 |
| GTCCT | 1546485 | 1.0371544 | 18.721283 | 7 |
| ACCGA | 1442755 | 1.0218822 | 5.4850507 | 70-74 |
| CCTAA | 1553410 | 1.0193509 | 5.7916875 | 25-29 |
| AGGCT | 1751710 | 1.0190016 | 5.182817 | 50-54 |
| GTACT | 1881810 | 1.0141865 | 9.032861 | 4 |
| GAGGT | 2044530 | 1.0038432 | 5.08624 | 35-39 |
| ACCGT | 1446785 | 0.9971419 | 19.219885 | 4 |
| TGGCA | 1708100 | 0.99363303 | 5.107209 | 65-69 |
| GGTAC | 1692240 | 0.9844069 | 9.649413 | 3 |
| TCCTA | 1539375 | 0.98293954 | 17.80358 | 8 |
| TGATC | 1818185 | 0.97989637 | 7.6897645 | 9 |
| CCGTC | 1111955 | 0.9536649 | 23.570236 | 5 |
| CAATT | 1856185 | 0.95246285 | 13.471968 | 4 |
| CCGAT | 1362230 | 0.9388656 | 5.6230326 | 75-79 |
| CTCCC | 920780 | 0.93563133 | 6.0977273 | 3 |
| GGCTA | 1594765 | 0.927704 | 5.3400817 | 55-59 |
| ACCTC | 1134800 | 0.92664325 | 10.558414 | 4 |
| GGTCA | 1588170 | 0.9238675 | 15.333469 | 9 |
| CCCGC | 825885 | 0.90581405 | 5.5125012 | 25-29 |
| CTGAC | 1313515 | 0.9052906 | 6.2979407 | 1 |
| GTACC | 1310350 | 0.90310913 | 5.191989 | 85-89 |
| CGAGT | 1546170 | 0.8994353 | 23.274294 | 2 |
| CAGAC | 1266785 | 0.89724517 | 9.632285 | 1 |
| TCCCC | 876785 | 0.8909267 | 5.718436 | 4 |
| CGGGC | 1120325 | 0.87535244 | 31.555286 | 7 |
| CGGTA | 1502795 | 0.8742033 | 5.6320677 | 5 |
| CGGGG | 1324580 | 0.87352806 | 7.6688137 | 6 |
| AATTG | 2015000 | 0.87269306 | 10.80812 | 5 |
| CCTCT | 1089805 | 0.865938 | 10.446272 | 5 |
| CACGG | 1162200 | 0.86457855 | 7.7022524 | 4 |
| GTCGG | 1411840 | 0.862608 | 24.937805 | 5 |
| CGTCC | 996825 | 0.8549241 | 23.3134 | 6 |
| TGGCC | 1171295 | 0.8478804 | 5.8121705 | 1 |
| TCCGC | 983255 | 0.84328574 | 5.1343694 | 65-69 |
| GCAAC | 1189505 | 0.8425089 | 5.1959124 | 1 |
| AGTCG | 1430370 | 0.8320723 | 23.472399 | 4 |
| TGCCA | 1199350 | 0.8266067 | 6.3440495 | 1 |
| CGGTT | 1417395 | 0.8023214 | 6.0761366 | 4 |
| TAGCG | 1373240 | 0.79883873 | 5.0527363 | 75-79 |
| CCGGT | 1087580 | 0.78728056 | 8.07701 | 3 |
| GGTTG | 1629640 | 0.77859014 | 6.785145 | 5 |
| CTCCG | 876960 | 0.7521221 | 5.102668 | 65-69 |
| CTGAT | 1355080 | 0.73030967 | 6.901532 | 8 |
| ATACC | 1095330 | 0.7187579 | 18.244663 | 2 |
| CCCGG | 720810 | 0.6672675 | 9.722884 | 2 |
| GACCT | 941885 | 0.64915866 | 8.779016 | 3 |
| GATAC | 1101710 | 0.6101894 | 15.312113 | 1 |