| Sequence |
Count |
Percentage |
Possible Source |
| GCGAGTCGGGCTGTTTGGGAATGCAGCCCTAAGTGGGAGGTAAATTCCTT |
226275 |
1.5899012480030938 |
No Hit |
| ACCCAATTGGTCAAGGAAGCTTCTCTGACGGTATGCCTTTAGGAATCTCT |
199424 |
1.4012350744968247 |
No Hit |
| GATACCGTCCTAGTCTCAACCATAAACGATGCCGACTAGGGATTGGCGGA |
139092 |
0.9773176196541656 |
No Hit |
| ACATGGGTCGTGAGTGGGAACTTAGCTACCGTTTAGGTATGCGTCCTTGG |
83065 |
0.5836488660496165 |
No Hit |
| GGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAAC |
77593 |
0.5452003426640329 |
No Hit |
| CTGGTACTTTCAACTTCATGATTGTGTTCCAAGCTGAACACAACATCCTT |
73428 |
0.5159353390271624 |
No Hit |
| CTGACGGTATGCCTTTAGGAATCTCTGGTACTTTCAACTTCATGATTGTG |
67917 |
0.47721278559551933 |
No Hit |
| ATGGTTCGGTGTTTTAATGATTCCTACTCTATTAACTGCAACTTCTGTAT |
67850 |
0.47674201602921185 |
No Hit |
| GCTACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGGAAGCTTCTCTGA |
64952 |
0.45637947568355736 |
No Hit |
| GCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTCTGCGA |
64892 |
0.45595789099731204 |
No Hit |
| GACTGCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTCT |
61205 |
0.43005151202753006 |
No Hit |
| CATGGAGAGTTTGATCCTGGCTCAGGATGAACGCTGGCGGCATGCTTAAC |
55771 |
0.39186999227656855 |
No Hit |
| CAGACCTCTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAA |
49215 |
0.34580483889281743 |
No Hit |
| GTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAACTTC |
46548 |
0.3270653995892079 |
No Hit |
| GTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTT |
44381 |
0.3118391660043103 |
No Hit |
| GCTTGGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGCACTAT |
44032 |
0.3093869484126493 |
No Hit |
| CAACACGGGGAAACTTACCAGGTCCAGACATAGTAAGGATTGACAGATTG |
38911 |
0.2734046954416015 |
No Hit |
| TGCTACATGGGTCGTGAGTGGGAACTTAGCTACCGTTTAGGTATGCGTCC |
37745 |
0.26521189970556525 |
No Hit |
| CCCCGGTTGTTACCCGACTACCAATAGGGGTGCTTCGTGCACCCCCAGAA |
37667 |
0.2646638396134462 |
No Hit |
| TGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTT |
37274 |
0.2619024599185386 |
No Hit |
| AAGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCGCC |
36656 |
0.25756013765021063 |
No Hit |
| AGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCGCCT |
36305 |
0.25509386723567484 |
No Hit |
| GGTACTTTCAACTTCATGATTGTGTTCCAAGCTGAACACAACATCCTTAT |
35921 |
0.25239572524370407 |
No Hit |
| CAAACGAATAAGGGCTTATGGTGGATACCTAGGCACCCAGAGACGAGGAA |
35658 |
0.25054777903566156 |
No Hit |
| CCTGTTGCGGCTGCTACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGG |
35337 |
0.24829230096424848 |
No Hit |
| TGCCAGTAGTCATATGCTTGTCTCAAAGATTAAGCCATGCATGTGTAAGT |
35248 |
0.24766695034631778 |
No Hit |
| TGGGTCGGGGGGAGCGGTCCGCCCCTTGTGGGTGTGCACTGGTCCACTCG |
34843 |
0.24482125371416108 |
No Hit |
| AGGCACCCAGAGACGAGGAAGGGCGTAGCAAGCGACGAAATGCTTCGGGG |
34300 |
0.2410059123036399 |
No Hit |
| GCAACTTCTGTATTTATTATTGCTTTCATTGCAGCTCCTCCTGTAGATAT |
33589 |
0.23601013377163152 |
No Hit |
| GATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAAGCGGAGGAG |
33408 |
0.23473835330145779 |
No Hit |
| TGACTGCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTC |
30955 |
0.21750256604545695 |
No Hit |
| ACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTCGCTTCTGCGACTG |
30600 |
0.21500818998517146 |
No Hit |
| GGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTC |
30270 |
0.21268947421082157 |
No Hit |
| ACCTCCCCGGTTGTTACCCGACTACCAATAGGGGTGCTTCGTGCACCCCC |
30056 |
0.21118582216321288 |
No Hit |
| CCTGTAGATATTGATGGTATCCGTGAGCCTGTTTCTGGTTCTCTTCTTTA |
29673 |
0.20849470658267952 |
No Hit |
| AGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAG |
28256 |
0.19853828157584985 |
No Hit |
| GACGGTATGCCTTTAGGAATCTCTGGTACTTTCAACTTCATGATTGTGTT |
28002 |
0.19675357307074415 |
No Hit |
| CAAGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCGC |
26094 |
0.18334718004813935 |
No Hit |
| CCACATGCTTGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTA |
24870 |
0.1747468524487325 |
No Hit |
| GATGGTTCGGTGTTTTAATGATTCCTACTCTATTAACTGCAACTTCTGTA |
24250 |
0.17039047735752966 |
No Hit |
| CTGATCAGGAAAGACTACCCGCTGAGTTTAAGCATATCACTAAGCGGAGG |
23437 |
0.16467800485890405 |
No Hit |
| CTATTCAGTGCTATGCATGGTTCCTTAGTAACTTCAAGTTTAATCCGTGA |
22003 |
0.15460213085763816 |
No Hit |
| ATGCATGGTTCCTTAGTAACTTCAAGTTTAATCCGTGAAACTACTGAGAA |
21856 |
0.15356924837633684 |
No Hit |
| GAACACAACATCCTTATGCACCCATTCCACATGCTTGGTGTAGCTGGTGT |
21652 |
0.15213586044310237 |
No Hit |
| GGTTCGGTGTTTTAATGATTCCTACTCTATTAACTGCAACTTCTGTATTT |
21541 |
0.1513559287735483 |
No Hit |
| ACATGCTTGGTGTAGCTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATG |
21528 |
0.1512645854248618 |
No Hit |
| AAGTGGGAGGTAAATTCCTTCTAAGGCTAAATATTGGCAAGAGACCGATA |
21048 |
0.14789190793489834 |
No Hit |
| GAAACATCTTAGTAGCCAGAGGAAAAGAAAGCAAAAGCGATTCCCATAGT |
20644 |
0.14505323771417908 |
No Hit |
| GTATTTATTATTGCTTTCATTGCAGCTCCTCCTGTAGATATTGATGGTAT |
20602 |
0.14475812843380728 |
No Hit |
| GCAAGCCTATGGGGTCGCTTCTGCGACTGGGTTACCAGCACTGAAAACCG |
20412 |
0.1434231102606967 |
No Hit |
| TGGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGCACTATGGC |
20062 |
0.14096386625759835 |
No Hit |
| GACACTGAGAGACGAAAGCTAGGGGAGCGAATGGGATTAGATACCCCAGT |
19747 |
0.13875054665480985 |
No Hit |
| ATTAACTGCAACTTCTGTATTTATTATTGCTTTCATTGCAGCTCCTCCTG |
19420 |
0.1364529101147722 |
No Hit |
| CCACGAGGCGCGATCTGCGAGTCGGGCTGTTTGGGAATGCAGCCCTAAGT |
19418 |
0.13643885729189736 |
No Hit |
| TTACATCGGATGGTTCGGTGTTTTAATGATTCCTACTCTATTAACTGCAA |
18694 |
0.13135173541120246 |
No Hit |
| GGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTAACTTCAAGTTTAAT |
18547 |
0.13031885292990117 |
No Hit |
| TACCATGACTGCTACTTTAGAAAGACGCGAAAGCGCAAGCCTATGGGGTC |
18243 |
0.1281828238529243 |
No Hit |
| CAACGCGAAGAACCTTACCAGGGCTTGACATGCCGCGAATCCTTTTGAAA |
18214 |
0.12797905792123898 |
No Hit |
| TGGCAAGAGACCGATAGCGAACAAGTACCGCGAGGGAAAGATGAAAAGGA |
18012 |
0.12655972281087935 |
No Hit |
| ACATCGGTAGGGGAGCGTTCCGCCTTAGAGGGAAGCATCAGCGCGAGCAG |
17946 |
0.12609597965600938 |
No Hit |
| ACCAATAGGGGTGCTTCGTGCACCCCCAGAAGCATGTGTCGAGAGCTCCC |
17932 |
0.12599760989588543 |
No Hit |
| GCGACTGGGTTACCAGCACTGAAAACCGCCTTTACATCGGATGGTTCGGT |
17821 |
0.1252176782263314 |
No Hit |
| CCTCCTGTAGATATTGATGGTATCCGTGAGCCTGTTTCTGGTTCTCTTCT |
17726 |
0.12455016913977612 |
No Hit |
| AACACGGACCAAGGAGTCTAACATGTATGCAAGCAGGTGGGCGGCAAACC |
17556 |
0.12335567919541406 |
No Hit |
| GCTCACGGTTACTTTGGTAGATTAATTTTCCAATACGCTAGCTTTAACAA |
17512 |
0.12304651709216739 |
No Hit |
| CTGGTGTATTCGGTGGCTCTCTATTCAGTGCTATGCATGGTTCCTTAGTA |
17299 |
0.12154989145599611 |
No Hit |
| CCACGTAGTTGGTTAGCTACTTCTCACTTTGTATTAGGATTTTTCTTCTT |
17070 |
0.11994084323682605 |
No Hit |
| TTCATGCGTTGGAGAGACCGTTTCTTATTCTGTGCAGAAGCTATTTACAA |
17066 |
0.11991273759107633 |
No Hit |
| GCAACACGGGGAAACTTACCAGGTCCAGACATAGTAAGGATTGACAGATT |
16295 |
0.11449537437282252 |
No Hit |
| GTAGGTGGCTTTTTAAGTCTGCTGTCAAATCCCAGGGCTCAACCCTGGAC |
16243 |
0.11413000097807648 |
No Hit |
| GCAGCTATCGGTTTGCACTTCTACCCAATTTGGGAAGCTGCTTCCGTTGA |
15837 |
0.11127727793448236 |
No Hit |
| ATCACTAAGCGGAGGAGAAGAAACTAACCAGGATTTCCCTAGTAACGGCG |
15622 |
0.10976659947543622 |
No Hit |
| GCTAAAAACTATGGTAGAGCTGTATACGAATGTCTTCGTGGTGGACTTGA |
15509 |
0.10897261498300734 |
No Hit |
| TACCAGCACTGAAAACCGCCTTTACATCGGATGGTTCGGTGTTTTAATGA |
15257 |
0.10720195930077651 |
No Hit |
| GGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGCACTATGGCT |
14982 |
0.10526969615548493 |
No Hit |
| TAGTAGCCAGAGGAAAAGAAAGCAAAAGCGATTCCCATAGTAGCGGCGAG |
14947 |
0.10502377175517509 |
No Hit |
| AAGCGGAGGAGAAGAAACTAACCAGGATTTCCCTAGTAACGGCGAGCGAA |
14938 |
0.10496053405223828 |
No Hit |
| ACCTGGTTGATCCTGCCAGTAGTCATATGCTTGTCTCAAAGATTAAGCCA |
14931 |
0.10491134917217632 |
No Hit |
| GTGAGCCTGTTTCTGGTTCTCTTCTTTACGGAAACAACATCATCTCTGCT |
14887 |
0.10460218706892965 |
No Hit |
| GGGTTACCAGCACTGAAAACCGCCTTTACATCGGATGGTTCGGTGTTTTA |
14772 |
0.1037941497536259 |
No Hit |
| GTTGCGGCTGCTACTGCTGTTTTCTTGATCTACCCAATTGGTCAAGGAAG |
14720 |
0.10342877635887986 |
No Hit |
| TAGCTGCTTGGCCTGTAGTAGGTATCTGGTTCACTGCATTAGGTATCAGC |
14447 |
0.10151056603646313 |
No Hit |
| ATACACTTGGGTAACTTATTATTATTTTACAAACCAAGATTTACCATGAC |
14402 |
0.10119437752177908 |
No Hit |
| GCCCTAGGTAACTTCTGTATTATAGTTGCTCCAGTTTTAAACTGAAAAAA |
14326 |
0.10066037025253484 |
No Hit |
![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality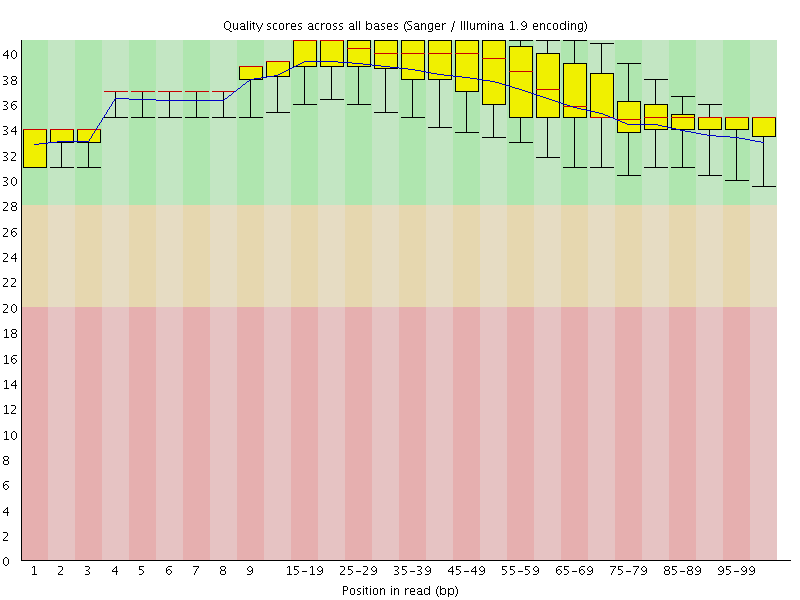
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores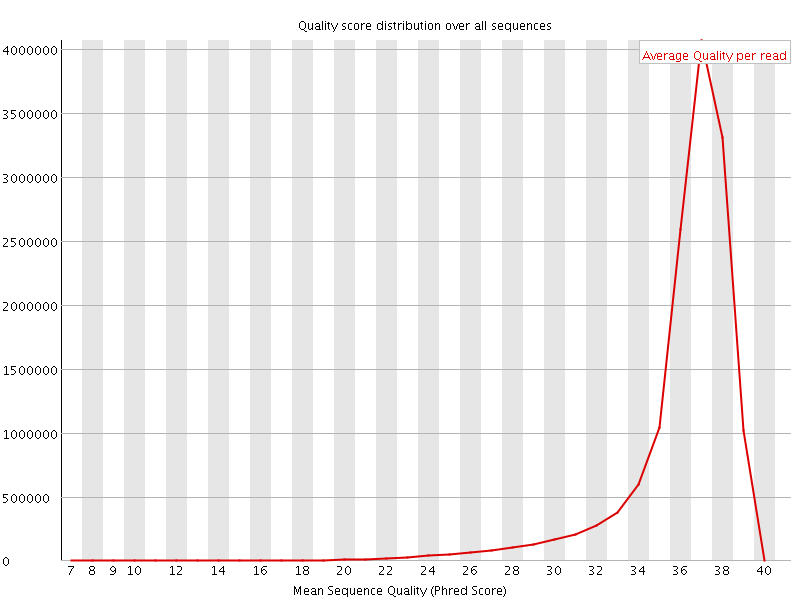
![[OK]](Icons/tick.png) Per base sequence content
Per base sequence content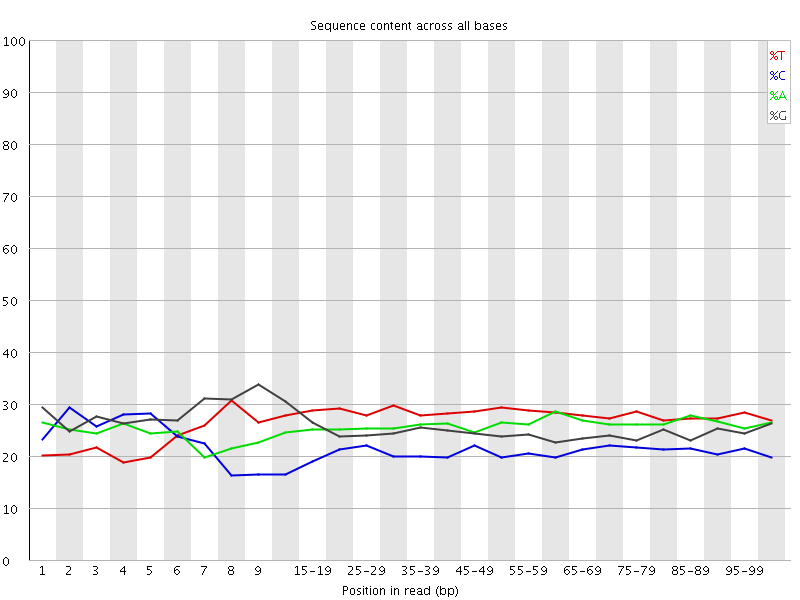
![[WARN]](Icons/warning.png) Per base GC content
Per base GC content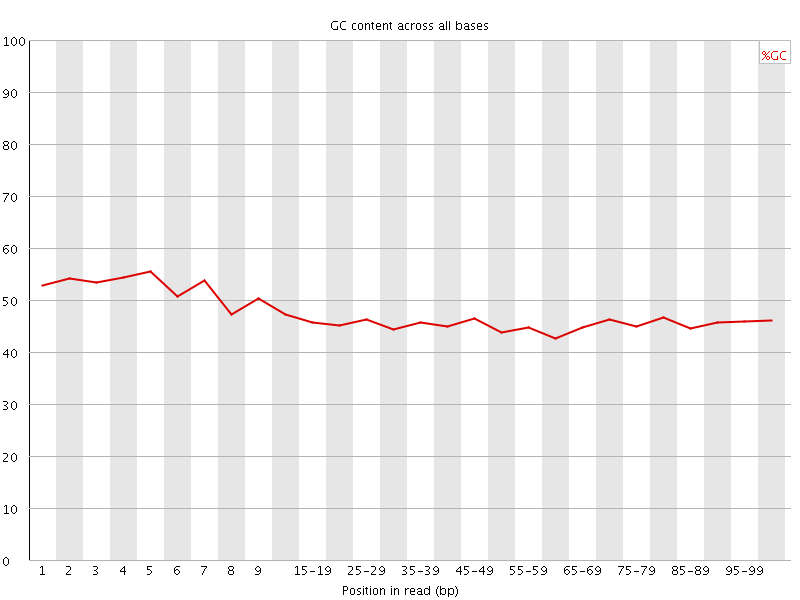
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content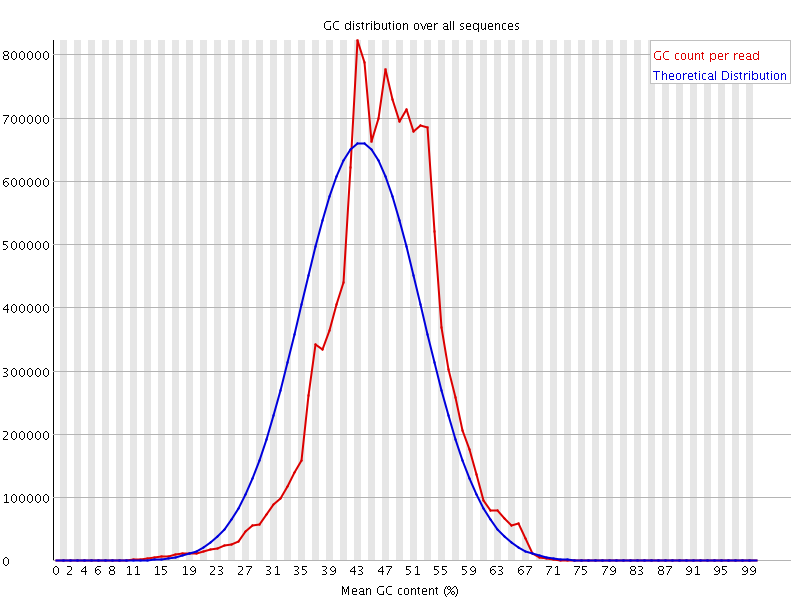
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution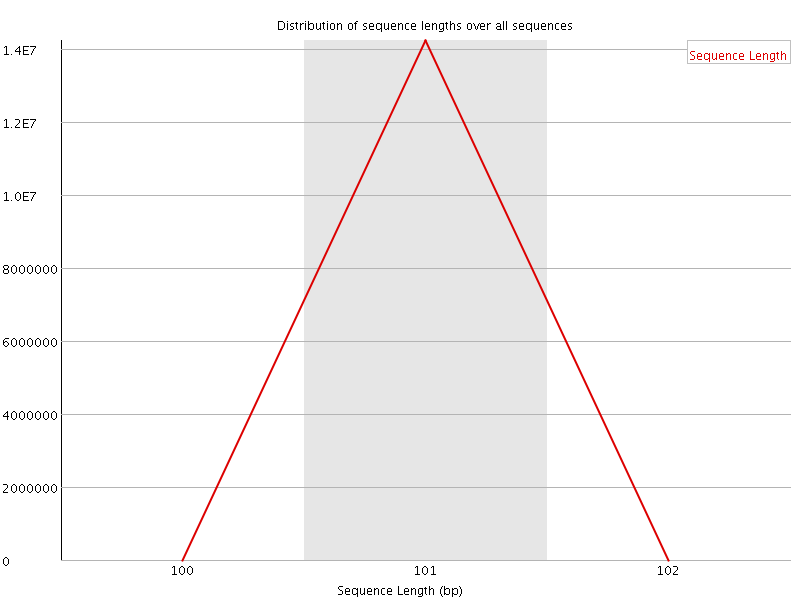
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels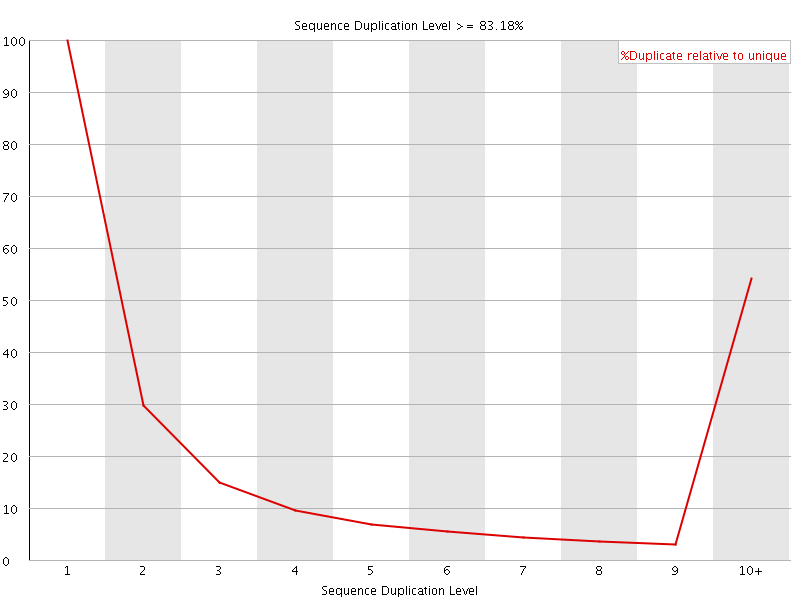
![[FAIL]](Icons/error.png) Overrepresented sequences
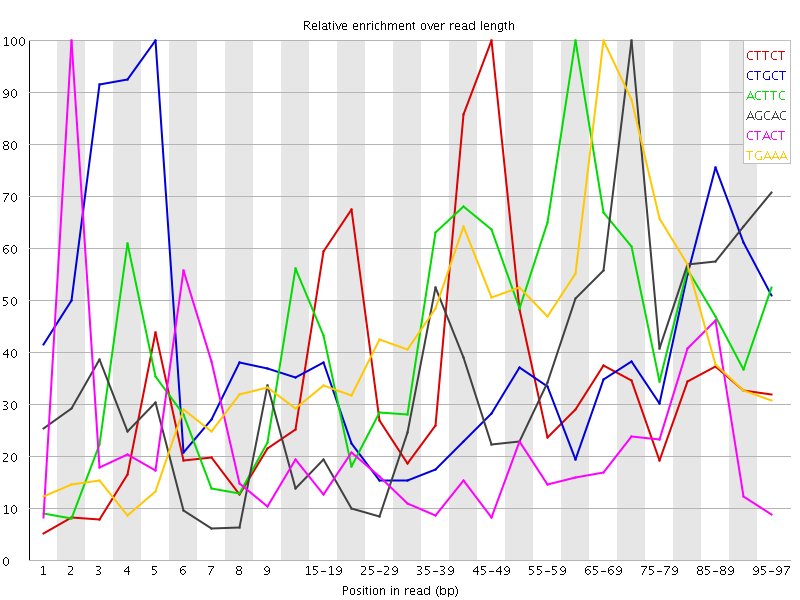
Overrepresented sequences![[WARN]](Icons/warning.png) Kmer Content
Kmer Content